当前位置:
X-MOL 学术
›
Lett. Appl. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
2019_nCoV: Rapid classification of betacoronaviruses and identification of traditional Chinese medicine as potential origin of zoonotic coronaviruses
Letters in Applied Microbiology ( IF 2.4 ) Pub Date : 2020-05-01 , DOI: 10.1111/lam.13285 T M Wassenaar 1 , Y Zou 2
Letters in Applied Microbiology ( IF 2.4 ) Pub Date : 2020-05-01 , DOI: 10.1111/lam.13285 T M Wassenaar 1 , Y Zou 2
Affiliation
|
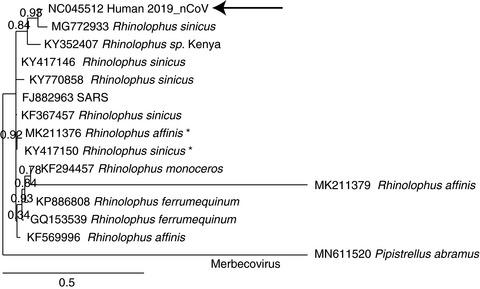
The current outbreak of a novel severe acute respiratory syndrome‐like coronavirus, 2019_nCoV (now named SARS‐CoV‐2), illustrated difficulties in identifying a novel coronavirus and its natural host, as the coding sequences of various Betacoronavirus species can be highly diverse. By means of whole‐genome sequence comparisons, we demonstrate that the noncoding flanks of the viral genome can be used to correctly separate the recognized four betacoronavirus subspecies. The conservation would be sufficient to define target sequences that could, in theory, classify novel virus species into their subspecies. Only 253 upstream noncoding sequences of Sarbecovirus are sufficient to identify genetic similarities between species of this subgenus. Furthermore, it was investigated which bat species have commercial value in China, and would thus likely be handled for trading purposes. A number of coronavirus genomes have been published that were obtained from such bat species. These bats are used in Traditional Chinese Medicine, and their handling poses a potential risk to cause zoonotic coronavirus epidemics.
更新日期:2020-05-01


























 京公网安备 11010802027423号
京公网安备 11010802027423号