当前位置:
X-MOL 学术
›
RSC Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Discovery of a novel kinase hinge binder fragment by dynamic undocking
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2020-03-04 , DOI: 10.1039/c9md00519f Moira Rachman 1, 2, 3, 4, 5 , Dávid Bajusz 5, 6, 7, 8 , Anasztázia Hetényi 8, 9, 10, 11 , Andrea Scarpino 5, 6, 7, 8 , Balázs Merő 6, 7, 8, 12 , Attila Egyed 5, 6, 7, 8 , László Buday 6, 7, 8, 12 , Xavier Barril 1, 2, 3, 4, 13 , György M. Keserű 5, 6, 7, 8
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2020-03-04 , DOI: 10.1039/c9md00519f Moira Rachman 1, 2, 3, 4, 5 , Dávid Bajusz 5, 6, 7, 8 , Anasztázia Hetényi 8, 9, 10, 11 , Andrea Scarpino 5, 6, 7, 8 , Balázs Merő 6, 7, 8, 12 , Attila Egyed 5, 6, 7, 8 , László Buday 6, 7, 8, 12 , Xavier Barril 1, 2, 3, 4, 13 , György M. Keserű 5, 6, 7, 8
Affiliation
|
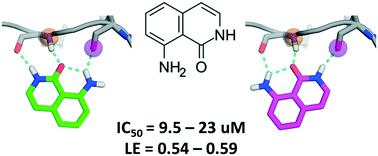
One of the key motifs of type I kinase inhibitors is their interactions with the hinge region of ATP binding sites. These interactions contribute significantly to the potency of the inhibitors; however, only a tiny fraction of the available chemical space has been explored with kinase inhibitors reported in the last twenty years. This paper describes a workflow utilizing docking with rDock and dynamic undocking (DUck) for the virtual screening of fragment libraries in order to identify fragments that bind to the kinase hinge region. We have identified 8-amino-2H-isoquinolin-1-one (MR1), a novel and potent hinge binding fragment, which was experimentally tested on a diverse set of kinases, and is hereby suggested for future fragment growing or merging efforts against various kinases, particularly MELK. Direct binding of MR1 to MELK was confirmed by STD-NMR, and its binding to the ATP-pocket was confirmed by a new competitive binding assay based on microscale thermophoresis.
中文翻译:

通过动态脱壳发现新型激酶铰链结合剂片段
I型激酶抑制剂的关键基元之一是它们与ATP结合位点的铰链区的相互作用。这些相互作用极大地抑制了抑制剂的效力。然而,在过去的二十年中,激酶抑制剂仅探索了可利用化学空间的一小部分。本文介绍了一种利用rDock对接和动态对接(DUck)对片段库进行虚拟筛选的工作流程,以识别与激酶铰链区结合的片段。我们已经确定了8-amino-2 H -isoquinolin-1-one(MR1),这是一种新颖而有效的铰链结合片段,已在多种激酶上进行了实验测试,因此建议用于未来的片段增长或针对各种激酶(尤其是MELK)的合并努力。通过STD-NMR证实了MR1与MELK的直接结合,并通过基于微尺度热泳的新型竞争结合试验证实了MR1与ATP口袋的结合。
更新日期:2020-03-04
中文翻译:

通过动态脱壳发现新型激酶铰链结合剂片段
I型激酶抑制剂的关键基元之一是它们与ATP结合位点的铰链区的相互作用。这些相互作用极大地抑制了抑制剂的效力。然而,在过去的二十年中,激酶抑制剂仅探索了可利用化学空间的一小部分。本文介绍了一种利用rDock对接和动态对接(DUck)对片段库进行虚拟筛选的工作流程,以识别与激酶铰链区结合的片段。我们已经确定了8-amino-2 H -isoquinolin-1-one(MR1),这是一种新颖而有效的铰链结合片段,已在多种激酶上进行了实验测试,因此建议用于未来的片段增长或针对各种激酶(尤其是MELK)的合并努力。通过STD-NMR证实了MR1与MELK的直接结合,并通过基于微尺度热泳的新型竞争结合试验证实了MR1与ATP口袋的结合。


























 京公网安备 11010802027423号
京公网安备 11010802027423号