Nature Biotechnology ( IF 46.9 ) Pub Date : 2020-03-02 , DOI: 10.1038/s41587-020-0434-2 Rodrigo Villaseñor 1 , Ramon Pfaendler 1 , Christina Ambrosi 1, 2 , Stefan Butz 1, 2 , Sara Giuliani 1 , Elana Bryan 3 , Thomas W Sheahan 3 , Annika L Gable 2, 4 , Nina Schmolka 1 , Massimiliano Manzo 1, 2 , Joël Wirz 1 , Christian Feller 5 , Christian von Mering 4 , Ruedi Aebersold 5, 6 , Philipp Voigt 3 , Tuncay Baubec 1
|
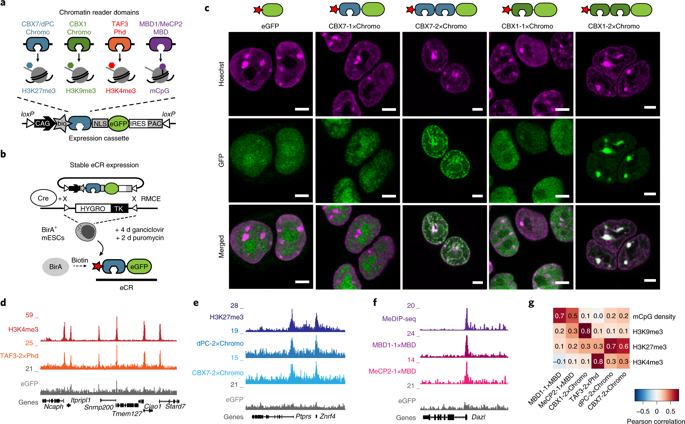
Chromatin modifications regulate genome function by recruiting proteins to the genome. However, the protein composition at distinct chromatin modifications has yet to be fully characterized. In this study, we used natural protein domains as modular building blocks to develop engineered chromatin readers (eCRs) selective for DNA methylation and histone tri-methylation at H3K4, H3K9 and H3K27 residues. We first demonstrated their utility as selective chromatin binders in living cells by stably expressing eCRs in mouse embryonic stem cells and measuring their subnuclear localization, genomic distribution and histone-modification-binding preference. By fusing eCRs to the biotin ligase BASU, we established ChromID, a method for identifying the chromatin-dependent protein interactome on the basis of proximity biotinylation, and applied it to distinct chromatin modifications in mouse stem cells. Using a synthetic dual-modification reader, we also uncovered the protein composition at bivalently modified promoters marked by H3K4me3 and H3K27me3. These results highlight the ability of ChromID to obtain a detailed view of protein interaction networks on chromatin.
中文翻译:

ChromID 识别染色质标记处的蛋白质相互作用组
染色质修饰通过向基因组募集蛋白质来调节基因组功能。然而,不同染色质修饰的蛋白质组成尚未完全表征。在这项研究中,我们使用天然蛋白质结构域作为模块化构建块来开发工程染色质阅读器 (eCR),该阅读器对 H3K4、H3K9 和 H3K27 残基的 DNA 甲基化和组蛋白三甲基化具有选择性。我们首先通过在小鼠胚胎干细胞中稳定表达 eCR 并测量它们的亚核定位、基因组分布和组蛋白修饰结合偏好,证明了它们在活细胞中作为选择性染色质结合剂的效用。通过将 eCR 与生物素连接酶 BASU 融合,我们建立了 ChromID,这是一种基于邻近生物素化识别染色质依赖性蛋白相互作用组的方法,并将其应用于小鼠干细胞中不同的染色质修饰。使用合成的双修饰阅读器,我们还发现了由 H3K4me3 和 H3K27me3 标记的二价修饰启动子的蛋白质组成。这些结果突出了 ChromID 获得染色质上蛋白质相互作用网络的详细视图的能力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号