JACC: Cardiovascular Interventions ( IF 11.3 ) Pub Date : 2020-03-02 , DOI: 10.1016/j.jcin.2020.01.219 Jean-Sébastien Hulot 1 , Bernard Chevalier 2 , Loic Belle 3 , Guillaume Cayla 4 , Khalife Khalife 5 , François Funck 6 , Romain Berthier 7 , Christophe Piot 8 , Muriel Tafflet 9 , Gilles Montalescot 10 ,
|
Objectives
The aim of this study was to evaluate prospectively the clinical impact of routine transmission of CYP2C19 genotype in the management of acute ST-segment elevation myocardial infarction with primary percutaneous coronary intervention.
Background
Response to clopidogrel differs widely among patients, notably because of CYP2C19 genetic polymorphisms.
Methods
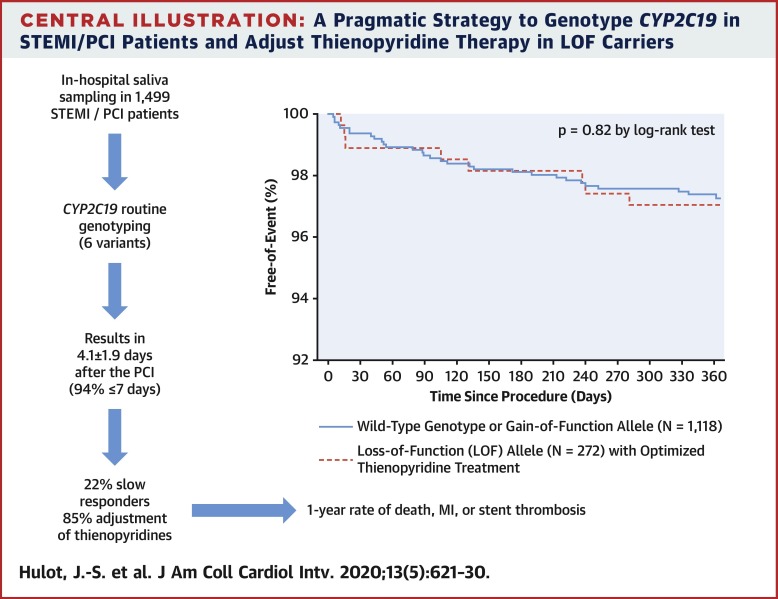
CYP2C19 genotype (6 alleles) was determined centrally and communicated within 4.1 ± 1.9 days of primary percutaneous coronary intervention in 1,445 patients with ST-segment elevation myocardial infarction recruited at 57 centers in France. CYP2C19 metabolic status was predicted from genotype and served to adjust thienopyridine treatment. The primary endpoint was differences in 12-month outcomes (death, myocardial infarction, and stent thrombosis) between patients with the wild-type genotype or gain-of-function allele (class 1, n = 1,118) and those with loss-of-function (LOF) alleles (class 2, n = 272) who received optimized thienopyridine treatment.
Results
Detection of LOF alleles resulted in adjustment of P2Y12 inhibition in 85% of patients, with significantly higher use of prasugrel or double-dose clopidogrel. The primary endpoint did not differ between class 1 and class 2 patients (3.31% vs. 3.04%, respectively; p = 0.82). In contrast, carriers of LOF alleles without treatment adjustment had significantly worse outcomes (15.6%; p < 0.05). Bleeding rates were not different between groups.
Conclusions
In a real-world setting, a complete CYPC2C19 genotype can be mostly determined in <7 days using analysis of saliva deoxyribonucleic acid collected during the in-hospital phase among patients with ST-segment elevation myocardial infarction treated with primary percutaneous coronary intervention. Genotype information led to stronger platelet inhibition treatment in the vast majority of LOF allele carriers and to similar clinical outcomes as in patients carrying the wild-type genotype or gain-of-function allele. (Genotyping Infarct Patients to Adjust and Normalize Thienopyridine Treatment [GIANT]; NCT01134380)
中文翻译:

常规CYP2C19基因分型以调整STEMI的主要PCI后的噻吩并吡啶治疗:GIANT研究的结果。
目标
这项研究的目的是前瞻性评估CYP2C19基因型常规传播对原发性经皮冠状动脉介入治疗急性ST段抬高型心肌梗塞的临床影响。
背景
患者之间对氯吡格雷的反应差异很大,特别是由于CYP2C19基因多态性。
方法
在法国的57个中心招募的1,445例ST段抬高型心肌梗死患者中,对CYP2C19基因型(6个等位基因)进行了中央确定,并在初次经皮冠状动脉介入治疗后4.1±1.9天内进行了沟通。从基因型预测CYP2C19的代谢状态,并用于调整噻吩并吡啶的治疗。主要终点是具有野生型基因型或功能获得性等位基因(1级,n = 1,118)与丧失功能的等位基因患者之间12个月结局(死亡,心肌梗塞和支架血栓形成)的差异。功能(LOF)等位基因(2类,n = 272),已接受优化的噻吩并吡啶治疗。
结果
LOF等位基因的检测可导致85%的患者调整P2Y 12抑制作用,而普拉格雷或双剂量氯吡格雷的使用率要高得多。1级和2级患者的主要终点没有差异(分别为3.31%和3.04%; p = 0.82)。相比之下,未经治疗调整的LOF等位基因携带者的结局明显较差(15.6%; p <0.05)。两组之间的出血率没有差异。
结论
在现实世界中,使用在原发性经皮冠状动脉介入治疗后ST段抬高型心肌梗死患者住院期间收集的唾液脱氧核糖核酸的分析,大部分CYPC2C19基因型可以在不到7天的时间内确定。基因型信息导致绝大多数LOF等位基因携带者接受更强的血小板抑制治疗,并产生与携带野生型基因型或功能获得等位基因的患者相似的临床结果。(对梗死患者进行基因分型以调整和规范噻吩并吡啶治疗[GIANT]; NCT01134380)


























 京公网安备 11010802027423号
京公网安备 11010802027423号