Acta Pharmaceutica Sinica B ( IF 14.5 ) Pub Date : 2020-02-27 , DOI: 10.1016/j.apsb.2020.02.008 Canrong Wu 1 , Yang Liu 2 , Yueying Yang 2 , Peng Zhang 2 , Wu Zhong 3 , Yali Wang 2 , Qiqi Wang 2 , Yang Xu 2 , Mingxue Li 2 , Xingzhou Li 3 , Mengzhu Zheng 1 , Lixia Chen 2 , Hua Li 1, 2
|
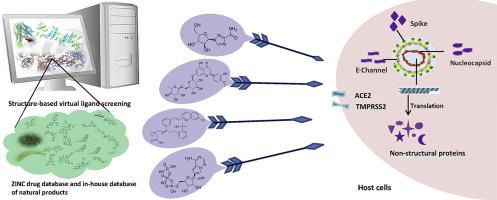
SARS-CoV-2 has caused tens of thousands of infections and more than one thousand deaths. There are currently no registered therapies for treating coronavirus infections. Because of time consuming process of new drug development, drug repositioning may be the only solution to the epidemic of sudden infectious diseases. We systematically analyzed all the proteins encoded by SARS-CoV-2 genes, compared them with proteins from other coronaviruses, predicted their structures, and built 19 structures that could be done by homology modeling. By performing target-based virtual ligand screening, a total of 21 targets (including two human targets) were screened against compound libraries including ZINC drug database and our own database of natural products. Structure and screening results of important targets such as 3-chymotrypsin-like protease (3CLpro), Spike, RNA-dependent RNA polymerase (RdRp), and papain like protease (PLpro) were discussed in detail. In addition, a database of 78 commonly used anti-viral drugs including those currently on the market and undergoing clinical trials for SARS-CoV-2 was constructed. Possible targets of these compounds and potential drugs acting on a certain target were predicted. This study will provide new lead compounds and targets for further in vitro and in vivo studies of SARS-CoV-2, new insights for those drugs currently ongoing clinical studies, and also possible new strategies for drug repositioning to treat SARS-CoV-2 infections.
中文翻译:

通过计算方法分析SARS-CoV-2的治疗靶点并发现潜在药物。
SARS-CoV-2造成了数以万计的感染,超过一千人死亡。目前尚无治疗冠状病毒感染的注册疗法。由于新药开发过程耗时,因此重新定位药物可能是解决突发性传染病流行的唯一方法。我们系统地分析了SARS-CoV-2基因编码的所有蛋白质,将它们与其他冠状病毒的蛋白质进行比较,预测了它们的结构,并构建了19个可以通过同源性建模完成的结构。通过执行基于靶标的虚拟配体筛选,针对包括ZINC药物数据库和我们自己的天然产物数据库在内的化合物库对总共21个靶标(包括两个人类靶标)进行了筛选。重要靶标的结构和筛选结果,例如3-胰凝乳蛋白酶样蛋白酶(3CLpro),详细讨论了穗,RNA依赖性RNA聚合酶(RdRp)和木瓜蛋白酶样蛋白酶(PLpro)。此外,还建立了包含78种常用抗病毒药物的数据库,包括目前市场上正在接受SARS-CoV-2临床试验的抗病毒药物。预测了这些化合物的可能目标以及作用于某个目标的潜在药物。这项研究将提供新的先导化合物和进一步的目标SARS-CoV-2的体外和体内研究,目前正在进行临床研究的那些药物的新见解,以及用于治疗SARS-CoV-2感染的药物重新定位的新策略。



























 京公网安备 11010802027423号
京公网安备 11010802027423号