npj Precision Oncology ( IF 7.9 ) Pub Date : 2020-02-24 , DOI: 10.1038/s41698-020-0109-y Tyler Landrith , Bing Li , Ashley A. Cass , Blair R. Conner , Holly LaDuca , Danielle B. McKenna , Kara N. Maxwell , Susan Domchek , Nichole A. Morman , Christopher Heinlen , Deborah Wham , Cathryn Koptiuch , Jennie Vagher , Ragene Rivera , Ann Bunnell , Gayle Patel , Jennifer L. Geurts , Morgan M. Depas , Shraddha Gaonkar , Sara Pirzadeh-Miller , Rebekah Krukenberg , Meredith Seidel , Robert Pilarski , Meagan Farmer , Khateriaa Pyrtel , Kara Milliron , John Lee , Elizabeth Hoodfar , Deepika Nathan , Amanda C. Ganzak , Sitao Wu , Huy Vuong , Dong Xu , Aarani Arulmoli , Melissa Parra , Lily Hoang , Bhuvan Molparia , Michele Fennessy , Susanne Fox , Sinead Charpentier , Julia Burdette , Tina Pesaran , Jessica Profato , Brandon Smith , Ginger Haynes , Emily Dalton , Joy Rae-Radecki Crandall , Ruth Baxter , Hsiao-Mei Lu , Brigette Tippin-Davis , Aaron Elliott , Elizabeth Chao , Rachid Karam
|
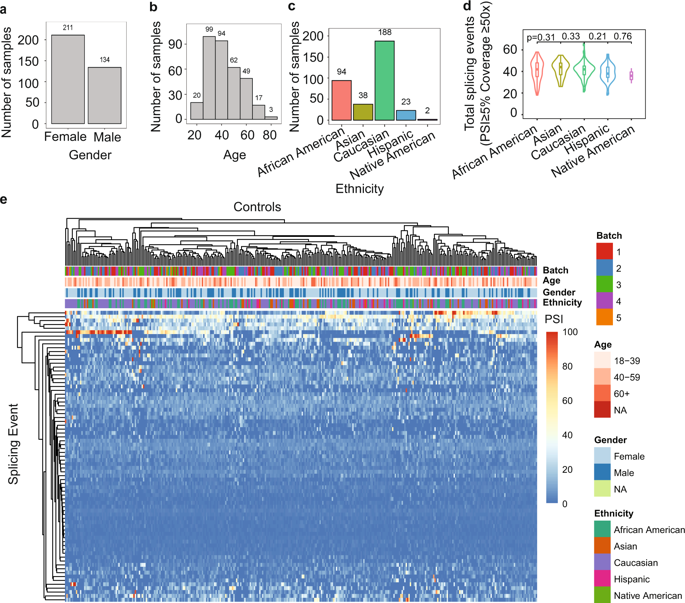
Germline variants in tumor suppressor genes (TSGs) can result in RNA mis-splicing and predisposition to cancer. However, identification of variants that impact splicing remains a challenge, contributing to a substantial proportion of patients with suspected hereditary cancer syndromes remaining without a molecular diagnosis. To address this, we used capture RNA-sequencing (RNA-seq) to generate a splicing profile of 18 TSGs (APC, ATM, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, MLH1, MSH2, MSH6, MUTYH, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, and TP53) in 345 whole-blood samples from healthy donors. We subsequently demonstrated that this approach can detect mis-splicing by comparing splicing profiles from the control dataset to profiles generated from whole blood of individuals previously identified with pathogenic germline splicing variants in these genes. To assess the utility of our TSG splicing profile to prospectively identify pathogenic splicing variants, we performed concurrent capture DNA and RNA-seq in a cohort of 1000 patients with suspected hereditary cancer syndromes. This approach improved the diagnostic yield in this cohort, resulting in a 9.1% relative increase in the detection of pathogenic variants, demonstrating the utility of performing simultaneous DNA and RNA genetic testing in a clinical context.
中文翻译:

通过捕获RNA-seq进行的剪接谱可鉴定肿瘤抑制基因中的致病种系变异
肿瘤抑制基因(TSGs)中的种系变异可能导致RNA错剪和易患癌症。然而,鉴定影响剪接的变体仍然是一个挑战,导致相当大比例的疑似遗传性癌症综合征患者没有进行分子诊断。为了解决这个问题,我们使用捕获RNA测序(RNA-seq)生成了18个TSG的剪接图谱(APC,ATM,BRCA1,BRCA2,BRIP1,CDH1,CHEK2,MLH1,MSH2,MSH6,MUTYH,NF1,PALB2,PMS2,PTEN,RAD51C,RAD51D和TP53)来自健康捐赠者的345个全血样本。我们随后证明了该方法可以通过比较对照数据集中的剪接图谱与先前从这些基因中的病原种系剪接变体鉴定出的个体全血生成的图谱来检测错接。为了评估我们的TSG剪接图谱用于前瞻性鉴定致病性剪接变体的实用性,我们在1000名疑似遗传性癌症综合征患者中进行了同时捕获DNA和RNA-seq。这种方法提高了该队列的诊断率,导致病原体变异的检测相对增加了9.1%,证明了在临床背景下同时进行DNA和RNA遗传测试的实用性。


























 京公网安备 11010802027423号
京公网安备 11010802027423号