当前位置:
X-MOL 学术
›
Adv. Funct. Mater.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Using SERS Tags to Image the Three‐Dimensional Structure of Complex Cell Models
Advanced Functional Materials ( IF 19.0 ) Pub Date : 2020-02-20 , DOI: 10.1002/adfm.201909655 Dorleta Jimenez de Aberasturi 1, 2 , Malou Henriksen‐Lacey 1 , Lucio Litti 3 , Judith Langer 1 , Luis M. Liz‐Marzán 1, 2
Advanced Functional Materials ( IF 19.0 ) Pub Date : 2020-02-20 , DOI: 10.1002/adfm.201909655 Dorleta Jimenez de Aberasturi 1, 2 , Malou Henriksen‐Lacey 1 , Lucio Litti 3 , Judith Langer 1 , Luis M. Liz‐Marzán 1, 2
Affiliation
|
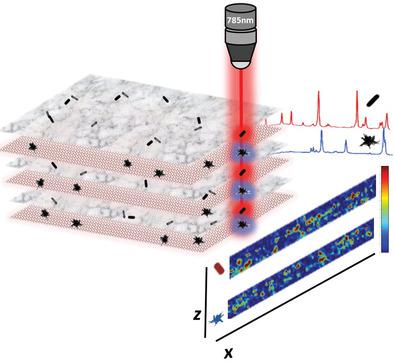
Methods to image complex 3D cell cultures are limited by issues such as fluorophore photobleaching and decomposition, poor excitation light penetration, and lack of complementary techniques to verify the 3D structure. Although it remains insufficiently demonstrated, surface‐enhanced Raman scattering (SERS) imaging is a promising tool for the characterization of biological complex systems. To this aim, a controllable 3D cell culture model which spans nearly 1 cm2 in surface footprint is designed. This structure is composed of fibroblasts containing SERS‐encoded nanoparticles (i.e., SERS tags), arranged in an alternating layered structure. This “sandwich” type structure allows monitoring of the SERS signals in the z‐axis and with mm dimensions in the xy‐axis. Taking advantage of correlative microscopy techniques such as electron microscopy, it is possible to corroborate nanoparticle positioning and distances in z‐depths of up to 150 µm. This study reveals a proof‐of‐concept method for detailed 3D SERS imaging of a complex, dense 3D cell culture model.
中文翻译:

使用SERS标签对复杂细胞模型的三维结构成像
对复杂的3D细胞培养物进行成像的方法受到诸如荧光团光漂白和分解,激发光穿透性差以及缺乏验证3D结构的互补技术等问题的限制。尽管仍然没有得到充分证明,但表面增强拉曼散射(SERS)成像是表征生物复杂系统的有前途的工具。为此,建立了一个可控的3D细胞培养模型,该模型跨越近1 cm 2设计了表面足迹。该结构由成纤维细胞组成,成纤维细胞含有SERS编码的纳米颗粒(即SERS标签),并以交替的分层结构排列。这种“三明治”型结构允许在z轴上监视SERS信号,在xy轴上监视mm尺寸。利用相关显微镜技术(例如电子显微镜),有可能在高达150 µm的z深度上证实纳米粒子的位置和距离。这项研究揭示了用于复杂,密集的3D细胞培养模型的详细3D SERS成像的概念验证方法。
更新日期:2020-04-06
中文翻译:

使用SERS标签对复杂细胞模型的三维结构成像
对复杂的3D细胞培养物进行成像的方法受到诸如荧光团光漂白和分解,激发光穿透性差以及缺乏验证3D结构的互补技术等问题的限制。尽管仍然没有得到充分证明,但表面增强拉曼散射(SERS)成像是表征生物复杂系统的有前途的工具。为此,建立了一个可控的3D细胞培养模型,该模型跨越近1 cm 2设计了表面足迹。该结构由成纤维细胞组成,成纤维细胞含有SERS编码的纳米颗粒(即SERS标签),并以交替的分层结构排列。这种“三明治”型结构允许在z轴上监视SERS信号,在xy轴上监视mm尺寸。利用相关显微镜技术(例如电子显微镜),有可能在高达150 µm的z深度上证实纳米粒子的位置和距离。这项研究揭示了用于复杂,密集的3D细胞培养模型的详细3D SERS成像的概念验证方法。


























 京公网安备 11010802027423号
京公网安备 11010802027423号