当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Atlas‐based GABA mapping with 3D MEGA‐MRSI: Cross‐correlation to single‐voxel MRS
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-02-20 , DOI: 10.1002/nbm.4275 Ruoyun E Ma 1, 2, 3 , James B Murdoch 4 , Wolfgang Bogner 5 , Ovidiu Andronesi 6 , Ulrike Dydak 2, 3
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-02-20 , DOI: 10.1002/nbm.4275 Ruoyun E Ma 1, 2, 3 , James B Murdoch 4 , Wolfgang Bogner 5 , Ovidiu Andronesi 6 , Ulrike Dydak 2, 3
Affiliation
|
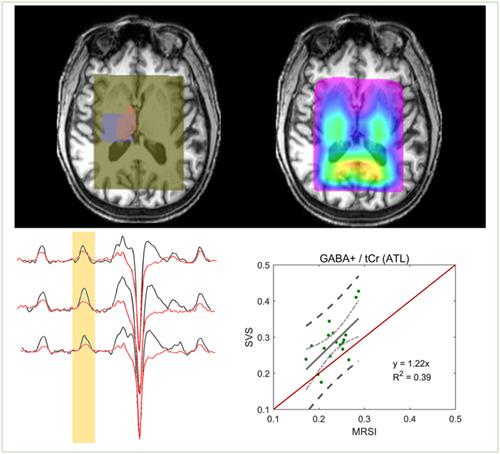
The purpose of this work is to develop and validate a new atlas‐based metabolite quantification pipeline for edited magnetic resonance spectroscopic imaging (MEGA‐MRSI) that enables group comparisons of brain structure‐specific GABA levels. By using brain structure masks segmented from high‐resolution MPRAGE images and coregistering these to MEGA‐LASER 3D MRSI data, an automated regional quantification of neurochemical levels is demonstrated for the example of the thalamus. Thalamic gamma‐aminobutyric acid + coedited macromolecules (GABA+) levels from 21 healthy subjects scanned at 3 T were cross‐validated both against a single‐voxel MEGA‐PRESS acquisition in the same subjects and same scan sessions, as well as alternative MRSI processing techniques (ROI approach, four‐voxel approach) using Pearson correlation analysis. In addition, reproducibility was compared across the MRSI processing techniques in test–retest data from 14 subjects. The atlas‐based approach showed a significant correlation with SV MEGA‐PRESS (correlation coefficient r [GABA+] = 0.63, P < 0.0001). However, the actual values for GABA+, NAA, tCr, GABA+/tCr and tNAA/tCr obtained from the atlas‐based approach showed an offset to SV MEGA‐PRESS levels, likely due to the fact that on average the thalamus mask used for the atlas‐based approach only occupied 30% of the SVS volume, ie, somewhat different anatomies were sampled. Furthermore, the new atlas‐based approach showed highly reproducible GABA+/tCr values with a low median coefficient of variance of 6.3%. In conclusion, the atlas‐based metabolite quantification approach enables a more brain structure‐specific comparison of GABA+ and other neurochemical levels across populations, even when using an MRSI technique with only cm‐level resolution. This approach was successfully cross‐validated against the typically used SVS technique as well as other different MRSI analysis methods, indicating the robustness of this quantification approach.
中文翻译:

基于图谱的 GABA 映射与 3D MEGA-MRSI:与单体素 MRS 的互相关
这项工作的目的是开发和验证一种新的基于图谱的代谢物量化管道,用于编辑磁共振波谱成像 (MEGA-MRSI),该管道能够对大脑结构特定的 GABA 水平进行组比较。通过使用从高分辨率 MPRAGE 图像中分割出来的大脑结构掩膜并将其与 MEGA-LASER 3D MRSI 数据进行配准,以丘脑为例,展示了神经化学水平的自动区域量化。21 名健康受试者在 3 T 下扫描的丘脑 γ-氨基丁酸 + 共编辑大分子 (GABA+) 水平针对相同受试者和相同扫描会话中的单体素 MEGA-PRESS 采集以及替代 MRSI 处理技术进行了交叉验证(ROI 方法,四体素方法)使用 Pearson 相关分析。此外,在 14 名受试者的重测数据中比较了 MRSI 处理技术的可重复性。基于图集的方法显示出与 SV MEGA-PRESS 的显着相关性(相关系数 r [GABA+] = 0.63,磷< 0.0001)。然而,从基于图集的方法获得的 GABA+、NAA、tCr、GABA+/tCr 和 tNAA/tCr 的实际值显示出与 SV MEGA-PRESS 水平的偏移,这可能是由于平均而言,用于基于图谱的方法仅占 SVS 体积的 30%,即采样的解剖结构略有不同。此外,新的基于图谱的方法显示出高度可重复的 GABA+/tCr 值,方差系数中位数仅为 6.3%。总之,基于图谱的代谢物量化方法能够对不同人群的 GABA+ 和其他神经化学水平进行更具大脑结构特异性的比较,即使使用只有厘米级分辨率的 MRSI 技术也是如此。这种方法成功地与常用的 SVS 技术以及其他不同的 MRSI 分析方法进行了交叉验证,
更新日期:2020-02-20
中文翻译:

基于图谱的 GABA 映射与 3D MEGA-MRSI:与单体素 MRS 的互相关
这项工作的目的是开发和验证一种新的基于图谱的代谢物量化管道,用于编辑磁共振波谱成像 (MEGA-MRSI),该管道能够对大脑结构特定的 GABA 水平进行组比较。通过使用从高分辨率 MPRAGE 图像中分割出来的大脑结构掩膜并将其与 MEGA-LASER 3D MRSI 数据进行配准,以丘脑为例,展示了神经化学水平的自动区域量化。21 名健康受试者在 3 T 下扫描的丘脑 γ-氨基丁酸 + 共编辑大分子 (GABA+) 水平针对相同受试者和相同扫描会话中的单体素 MEGA-PRESS 采集以及替代 MRSI 处理技术进行了交叉验证(ROI 方法,四体素方法)使用 Pearson 相关分析。此外,在 14 名受试者的重测数据中比较了 MRSI 处理技术的可重复性。基于图集的方法显示出与 SV MEGA-PRESS 的显着相关性(相关系数 r [GABA+] = 0.63,磷< 0.0001)。然而,从基于图集的方法获得的 GABA+、NAA、tCr、GABA+/tCr 和 tNAA/tCr 的实际值显示出与 SV MEGA-PRESS 水平的偏移,这可能是由于平均而言,用于基于图谱的方法仅占 SVS 体积的 30%,即采样的解剖结构略有不同。此外,新的基于图谱的方法显示出高度可重复的 GABA+/tCr 值,方差系数中位数仅为 6.3%。总之,基于图谱的代谢物量化方法能够对不同人群的 GABA+ 和其他神经化学水平进行更具大脑结构特异性的比较,即使使用只有厘米级分辨率的 MRSI 技术也是如此。这种方法成功地与常用的 SVS 技术以及其他不同的 MRSI 分析方法进行了交叉验证,



























 京公网安备 11010802027423号
京公网安备 11010802027423号