Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A gene-expression-based signature predicts survival in adults with T-cell lymphoblastic lymphoma: a multicenter study.
Leukemia ( IF 11.4 ) Pub Date : 2020-02-20 , DOI: 10.1038/s41375-020-0757-5 Xiao-Peng Tian 1, 2 , Dan Xie 1 , Wei-Juan Huang 3 , Shu-Yun Ma 1, 2 , Liang Wang 4, 5 , Yan-Hui Liu 6 , Xi Zhang 7 , Hui-Qiang Huang 2 , Tong-Yu Lin 2 , Hui-Lan Rao 8 , Mei Li 8 , Fang Liu 9 , Fen Zhang 6 , Li-Ye Zhong 10 , Li Liang 11 , Xiao-Liang Lan 12 , Juan Li 13 , Bing Liao 14 , Zhi-Hua Li 15 , Qiong-Lan Tang 15 , Qiong Liang 16 , Chun-Kui Shao 16 , Qiong-Li Zhai 17 , Run-Fen Cheng 17 , Qi Sun 18 , Kun Ru 17 , Xia Gu 19 , Xi-Na Lin 19 , Kun Yi 20 , Yue-Rong Shuang 21 , Xiao-Dong Chen 22 , Wei Dong 23 , Wei Sang 24 , Cai Sun 25 , Hui Liu 25 , Zhi-Gang Zhu 26 , Jun Rao 7 , Qiao-Nan Guo 27 , Ying Zhou 28 , Xiang-Ling Meng 29 , Yong Zhu 30 , Chang-Lu Hu 31 , Yi-Rong Jiang 32 , Ying Zhang 33 , Hong-Yi Gao 34 , Wen-Jun He 35 , Zhong-Jun Xia 36 , Xue-Yi Pan 37 , Hai Lan 38 , Guo-Wei Li 39 , Lu Liu 40 , Hui-Zheng Bao 40 , Li-Yan Song 3 , Tie-Bang Kang 1 , Qing-Qing Cai 1, 2
Leukemia ( IF 11.4 ) Pub Date : 2020-02-20 , DOI: 10.1038/s41375-020-0757-5 Xiao-Peng Tian 1, 2 , Dan Xie 1 , Wei-Juan Huang 3 , Shu-Yun Ma 1, 2 , Liang Wang 4, 5 , Yan-Hui Liu 6 , Xi Zhang 7 , Hui-Qiang Huang 2 , Tong-Yu Lin 2 , Hui-Lan Rao 8 , Mei Li 8 , Fang Liu 9 , Fen Zhang 6 , Li-Ye Zhong 10 , Li Liang 11 , Xiao-Liang Lan 12 , Juan Li 13 , Bing Liao 14 , Zhi-Hua Li 15 , Qiong-Lan Tang 15 , Qiong Liang 16 , Chun-Kui Shao 16 , Qiong-Li Zhai 17 , Run-Fen Cheng 17 , Qi Sun 18 , Kun Ru 17 , Xia Gu 19 , Xi-Na Lin 19 , Kun Yi 20 , Yue-Rong Shuang 21 , Xiao-Dong Chen 22 , Wei Dong 23 , Wei Sang 24 , Cai Sun 25 , Hui Liu 25 , Zhi-Gang Zhu 26 , Jun Rao 7 , Qiao-Nan Guo 27 , Ying Zhou 28 , Xiang-Ling Meng 29 , Yong Zhu 30 , Chang-Lu Hu 31 , Yi-Rong Jiang 32 , Ying Zhang 33 , Hong-Yi Gao 34 , Wen-Jun He 35 , Zhong-Jun Xia 36 , Xue-Yi Pan 37 , Hai Lan 38 , Guo-Wei Li 39 , Lu Liu 40 , Hui-Zheng Bao 40 , Li-Yan Song 3 , Tie-Bang Kang 1 , Qing-Qing Cai 1, 2
Affiliation
|
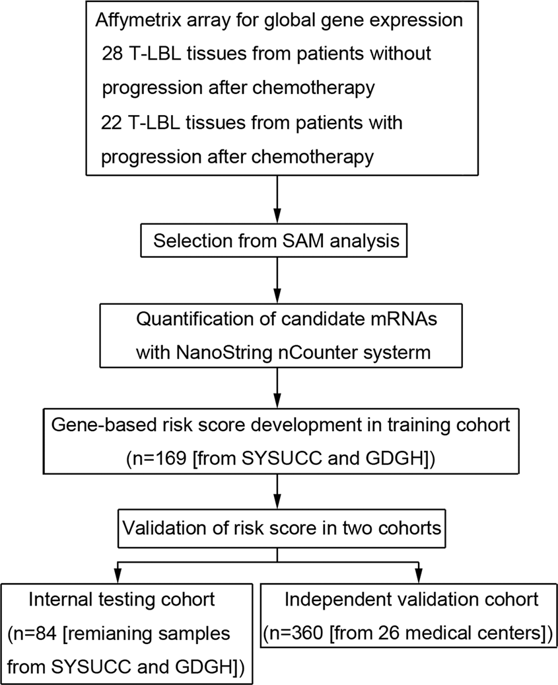
We aimed to establish a discriminative gene-expression-based classifier to predict survival outcomes of T-cell lymphoblastic lymphoma (T-LBL) patients. After exploring global gene-expression profiles of progressive (n = 22) vs. progression-free (n = 28) T-LBL patients, 43 differentially expressed mRNAs were identified. Then an eleven-gene-based classifier was established using LASSO Cox regression based on NanoString quantification. In the training cohort (n = 169), high-risk patients stratified using the classifier had significantly lower progression-free survival (PFS: hazards ratio 4.123, 95% CI 2.565-6.628; p < 0.001), disease-free survival (DFS: HR 3.148, 95% CI 1.857-5.339; p < 0.001), and overall survival (OS: HR 3.790, 95% CI 2.237-6.423; p < 0.001) compared with low-risk patients. The prognostic accuracy of the classifier was validated in the internal testing (n = 84) and independent validation cohorts (n = 360). A prognostic nomogram consisting of five independent variables including the classifier, lactate dehydrogenase levels, ECOG-PS, central nervous system involvement, and NOTCH1/FBXW7 status showed significantly greater prognostic accuracy than each single variable alone. The addition of a five-miRNA-based signature further enhanced the accuracy of this nomogram. Furthermore, patients with a nomogram score ≥154.2 significantly benefited from the BFM protocol. In conclusion, our nomogram comprising the 11-gene-based classifier may make contributions to individual prognosis prediction and treatment decision-making.
中文翻译:

一个基于基因表达的签名可以预测成人T细胞淋巴母细胞淋巴瘤的生存:一项多中心研究。
我们旨在建立一个基于判别性基因表达的分类器,以预测T细胞淋巴母细胞性淋巴瘤(T-LBL)患者的生存结果。在探究进行性(n = 22)与无进展(n = 28)T-LBL患者的整体基因表达谱后,鉴定出43种差异表达的mRNA。然后使用基于NanoString量化的LASSO Cox回归建立了基于11个基因的分类器。在训练队列中(n = 169),使用分类器进行分层的高危患者的无进展生存期明显降低(PFS:危险比4.123,95%CI 2.565-6.628; p <0.001),无病生存期(DFS) :与低危患者相比,HR 3.148,95%CI 1.857-5.339; p <0.001)和总体存活率(OS:HR 3.790,95%CI 2.237-6.423; p <0.001)。在内部测试(n = 84)和独立的验证队列(n = 360)中验证了分类器的预后准确性。由五个独立变量(包括分类器,乳酸脱氢酶水平,ECOG-PS,中枢神经系统受累和NOTCH1 / FBXW7状态)组成的预后诺模图显示的预后准确性要比单独每个单独变量高得多。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。由五个独立变量(包括分类器,乳酸脱氢酶水平,ECOG-PS,中枢神经系统受累和NOTCH1 / FBXW7状态)组成的预后诺模图显示的预后准确性要比单独每个单独变量高得多。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。由五个独立变量(包括分类器,乳酸脱氢酶水平,ECOG-PS,中枢神经系统受累和NOTCH1 / FBXW7状态)组成的预后诺模图显示的预后准确性要比单独每个单独变量高得多。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。和NOTCH1 / FBXW7状态显示的预后准确性明显高于每个单独的变量。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。和NOTCH1 / FBXW7状态显示的预后准确性明显高于每个单独的变量。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。
更新日期:2020-02-21
中文翻译:

一个基于基因表达的签名可以预测成人T细胞淋巴母细胞淋巴瘤的生存:一项多中心研究。
我们旨在建立一个基于判别性基因表达的分类器,以预测T细胞淋巴母细胞性淋巴瘤(T-LBL)患者的生存结果。在探究进行性(n = 22)与无进展(n = 28)T-LBL患者的整体基因表达谱后,鉴定出43种差异表达的mRNA。然后使用基于NanoString量化的LASSO Cox回归建立了基于11个基因的分类器。在训练队列中(n = 169),使用分类器进行分层的高危患者的无进展生存期明显降低(PFS:危险比4.123,95%CI 2.565-6.628; p <0.001),无病生存期(DFS) :与低危患者相比,HR 3.148,95%CI 1.857-5.339; p <0.001)和总体存活率(OS:HR 3.790,95%CI 2.237-6.423; p <0.001)。在内部测试(n = 84)和独立的验证队列(n = 360)中验证了分类器的预后准确性。由五个独立变量(包括分类器,乳酸脱氢酶水平,ECOG-PS,中枢神经系统受累和NOTCH1 / FBXW7状态)组成的预后诺模图显示的预后准确性要比单独每个单独变量高得多。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。由五个独立变量(包括分类器,乳酸脱氢酶水平,ECOG-PS,中枢神经系统受累和NOTCH1 / FBXW7状态)组成的预后诺模图显示的预后准确性要比单独每个单独变量高得多。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。由五个独立变量(包括分类器,乳酸脱氢酶水平,ECOG-PS,中枢神经系统受累和NOTCH1 / FBXW7状态)组成的预后诺模图显示的预后准确性要比单独每个单独变量高得多。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。和NOTCH1 / FBXW7状态显示的预后准确性明显高于每个单独的变量。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。和NOTCH1 / FBXW7状态显示的预后准确性明显高于每个单独的变量。添加基于五个miRNA的签名进一步提高了该列线图的准确性。此外,列线图评分≥154.2的患者从BFM方案中显着受益。总之,我们的诺模图包括基于11个基因的分类器,可能有助于个人预后的预测和治疗决策。



























 京公网安备 11010802027423号
京公网安备 11010802027423号