Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Deep Learning Approach to Antibiotic Discovery.
Cell ( IF 64.5 ) Pub Date : 2020-02-20 , DOI: 10.1016/j.cell.2020.01.021 Jonathan M Stokes 1 , Kevin Yang 2 , Kyle Swanson 2 , Wengong Jin 2 , Andres Cubillos-Ruiz 3 , Nina M Donghia 4 , Craig R MacNair 5 , Shawn French 5 , Lindsey A Carfrae 5 , Zohar Bloom-Ackermann 6 , Victoria M Tran 7 , Anush Chiappino-Pepe 8 , Ahmed H Badran 7 , Ian W Andrews 3 , Emma J Chory 9 , George M Church 10 , Eric D Brown 5 , Tommi S Jaakkola 2 , Regina Barzilay 11 , James J Collins 12
Cell ( IF 64.5 ) Pub Date : 2020-02-20 , DOI: 10.1016/j.cell.2020.01.021 Jonathan M Stokes 1 , Kevin Yang 2 , Kyle Swanson 2 , Wengong Jin 2 , Andres Cubillos-Ruiz 3 , Nina M Donghia 4 , Craig R MacNair 5 , Shawn French 5 , Lindsey A Carfrae 5 , Zohar Bloom-Ackermann 6 , Victoria M Tran 7 , Anush Chiappino-Pepe 8 , Ahmed H Badran 7 , Ian W Andrews 3 , Emma J Chory 9 , George M Church 10 , Eric D Brown 5 , Tommi S Jaakkola 2 , Regina Barzilay 11 , James J Collins 12
Affiliation
|
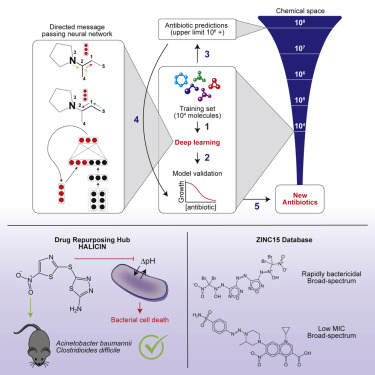
Due to the rapid emergence of antibiotic-resistant bacteria, there is a growing need to discover new antibiotics. To address this challenge, we trained a deep neural network capable of predicting molecules with antibacterial activity. We performed predictions on multiple chemical libraries and discovered a molecule from the Drug Repurposing Hub-halicin-that is structurally divergent from conventional antibiotics and displays bactericidal activity against a wide phylogenetic spectrum of pathogens including Mycobacterium tuberculosis and carbapenem-resistant Enterobacteriaceae. Halicin also effectively treated Clostridioides difficile and pan-resistant Acinetobacter baumannii infections in murine models. Additionally, from a discrete set of 23 empirically tested predictions from >107 million molecules curated from the ZINC15 database, our model identified eight antibacterial compounds that are structurally distant from known antibiotics. This work highlights the utility of deep learning approaches to expand our antibiotic arsenal through the discovery of structurally distinct antibacterial molecules.
中文翻译:

抗生素发现的深度学习方法。
由于耐抗生素细菌的迅速出现,人们越来越需要发现新的抗生素。为了应对这一挑战,我们训练了一个能够预测具有抗菌活性的分子的深度神经网络。我们对多个化学文库进行了预测,并从药物再利用中心 - halicin 中发现了一种分子,该分子在结构上与传统抗生素不同,并且对包括结核分枝杆菌和耐碳青霉烯类肠杆菌科在内的广泛的病原体系统发育谱显示出杀菌活性。Halicin 还有效治疗小鼠模型中的艰难梭菌和泛耐药鲍曼不动杆菌感染。此外,从来自 ZINC15 数据库的超过 1.07 亿个分子的 23 个经验测试预测的离散集合中,我们的模型确定了八种在结构上与已知抗生素相距甚远的抗菌化合物。这项工作突出了深度学习方法通过发现结构不同的抗菌分子来扩展我们的抗生素库的效用。
更新日期:2020-02-20
中文翻译:

抗生素发现的深度学习方法。
由于耐抗生素细菌的迅速出现,人们越来越需要发现新的抗生素。为了应对这一挑战,我们训练了一个能够预测具有抗菌活性的分子的深度神经网络。我们对多个化学文库进行了预测,并从药物再利用中心 - halicin 中发现了一种分子,该分子在结构上与传统抗生素不同,并且对包括结核分枝杆菌和耐碳青霉烯类肠杆菌科在内的广泛的病原体系统发育谱显示出杀菌活性。Halicin 还有效治疗小鼠模型中的艰难梭菌和泛耐药鲍曼不动杆菌感染。此外,从来自 ZINC15 数据库的超过 1.07 亿个分子的 23 个经验测试预测的离散集合中,我们的模型确定了八种在结构上与已知抗生素相距甚远的抗菌化合物。这项工作突出了深度学习方法通过发现结构不同的抗菌分子来扩展我们的抗生素库的效用。


























 京公网安备 11010802027423号
京公网安备 11010802027423号