当前位置:
X-MOL 学术
›
PLOS Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A molecular barcode to inform the geographical origin and transmission dynamics of Plasmodium vivax malaria.
PLOS Genetics ( IF 4.5 ) Pub Date : 2020-02-13 , DOI: 10.1371/journal.pgen.1008576 Ernest Diez Benavente 1 , Monica Campos 1 , Jody Phelan 1 , Debbie Nolder 1 , Jamille G Dombrowski 2 , Claudio R F Marinho 2 , Kanlaya Sriprawat 3 , Aimee R Taylor 4, 5 , James Watson 6, 7 , Cally Roper 1 , Francois Nosten 3, 6 , Colin J Sutherland 1 , Susana Campino 1 , Taane G Clark 1, 8
PLOS Genetics ( IF 4.5 ) Pub Date : 2020-02-13 , DOI: 10.1371/journal.pgen.1008576 Ernest Diez Benavente 1 , Monica Campos 1 , Jody Phelan 1 , Debbie Nolder 1 , Jamille G Dombrowski 2 , Claudio R F Marinho 2 , Kanlaya Sriprawat 3 , Aimee R Taylor 4, 5 , James Watson 6, 7 , Cally Roper 1 , Francois Nosten 3, 6 , Colin J Sutherland 1 , Susana Campino 1 , Taane G Clark 1, 8
Affiliation
|
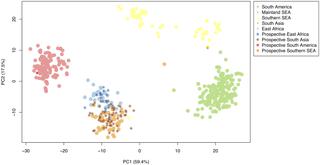
Although Plasmodium vivax parasites are the predominant cause of malaria outside of sub-Saharan Africa, they not always prioritised by elimination programmes. P. vivax is resilient and poses challenges through its ability to re-emerge from dormancy in the human liver. With observed growing drug-resistance and the increasing reports of life-threatening infections, new tools to inform elimination efforts are needed. In order to halt transmission, we need to better understand the dynamics of transmission, the movement of parasites, and the reservoirs of infection in order to design targeted interventions. The use of molecular genetics and epidemiology for tracking and studying malaria parasite populations has been applied successfully in P. falciparum species and here we sought to develop a molecular genetic tool for P. vivax. By assembling the largest set of P. vivax whole genome sequences (n = 433) spanning 17 countries, and applying a machine learning approach, we created a 71 SNP barcode with high predictive ability to identify geographic origin (91.4%). Further, due to the inclusion of markers for within population variability, the barcode may also distinguish local transmission networks. By using P. vivax data from a low-transmission setting in Malaysia, we demonstrate the potential ability to infer outbreak events. By characterising the barcoding SNP genotypes in P. vivax DNA sourced from UK travellers (n = 132) to ten malaria endemic countries predominantly not used in the barcode construction, we correctly predicted the geographic region of infection origin. Overall, the 71 SNP barcode outperforms previously published genotyping methods and when rolled-out within new portable platforms, is likely to be an invaluable tool for informing targeted interventions towards elimination of this resilient human malaria.
中文翻译:

分子条形码,可告知间日疟原虫疟疾的地理起源和传播动态。
尽管间日疟原虫寄生虫是撒哈拉以南非洲以外地区疟疾的主要诱因,但并非总是将消除疟疾方案作为其重点。间日疟原虫具有韧性,并通过其从人体肝脏休眠中重新出现的能力提出了挑战。随着观察到的耐药性增长和威胁生命的感染的报告不断增加,需要新的工具来为消除工作提供信息。为了阻止传播,我们需要更好地了解传播的动态,寄生虫的运动以及感染源,以设计针对性的干预措施。分子遗传学和流行病学用于追踪和研究疟疾寄生虫种群的方法已成功应用于恶性疟原虫物种,在这里我们寻求开发一种用于间日疟原虫的分子遗传工具。通过组装横跨17个国家/地区的最大的间日疟原虫全基因组序列(n = 433),并应用机器学习方法,我们创建了71个SNP条码,具有识别地理来源的高预测能力(91.4%)。此外,由于包括了针对群体变异性的标记,因此条形码还可以区分本地传输网络。通过使用马来西亚低传播环境下的间日疟原虫数据,我们证明了推断爆发事件的潜在能力。通过表征来自英国旅行者(n = 132)到十个疟疾流行国家(主要不用于条形码构建)的间日疟原虫DNA的条形码SNP基因型,我们正确地预测了感染起源的地理区域。总体,
更新日期:2020-03-05
中文翻译:

分子条形码,可告知间日疟原虫疟疾的地理起源和传播动态。
尽管间日疟原虫寄生虫是撒哈拉以南非洲以外地区疟疾的主要诱因,但并非总是将消除疟疾方案作为其重点。间日疟原虫具有韧性,并通过其从人体肝脏休眠中重新出现的能力提出了挑战。随着观察到的耐药性增长和威胁生命的感染的报告不断增加,需要新的工具来为消除工作提供信息。为了阻止传播,我们需要更好地了解传播的动态,寄生虫的运动以及感染源,以设计针对性的干预措施。分子遗传学和流行病学用于追踪和研究疟疾寄生虫种群的方法已成功应用于恶性疟原虫物种,在这里我们寻求开发一种用于间日疟原虫的分子遗传工具。通过组装横跨17个国家/地区的最大的间日疟原虫全基因组序列(n = 433),并应用机器学习方法,我们创建了71个SNP条码,具有识别地理来源的高预测能力(91.4%)。此外,由于包括了针对群体变异性的标记,因此条形码还可以区分本地传输网络。通过使用马来西亚低传播环境下的间日疟原虫数据,我们证明了推断爆发事件的潜在能力。通过表征来自英国旅行者(n = 132)到十个疟疾流行国家(主要不用于条形码构建)的间日疟原虫DNA的条形码SNP基因型,我们正确地预测了感染起源的地理区域。总体,


























 京公网安备 11010802027423号
京公网安备 11010802027423号