Nature Chemical Biology ( IF 14.8 ) Pub Date : 2020-02-10 , DOI: 10.1038/s41589-019-0458-4 Claire M Metrick 1, 2 , Emily A Peterson 3 , Joseph C Santoro 4 , Istvan J Enyedy 3 , Paramasivam Murugan 4 , TeYu Chen 3 , Klaus Michelsen 1 , Michael Cullivan 1 , Kerri A Spilker 1 , P Rajesh Kumar 1 , Tricia L May-Dracka 3 , Jayanth V Chodaparambil 1
|
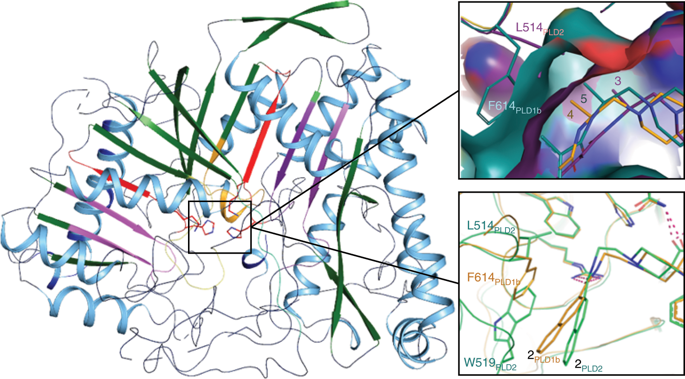
Phospholipase D enzymes (PLDs) are ubiquitous phosphodiesterases that produce phosphatidic acid (PA), a key second messenger and biosynthetic building block. Although an orthologous bacterial Streptomyces sp. strain PMF PLD structure was solved two decades ago, the molecular basis underlying the functions of the human PLD enzymes (hPLD) remained unclear based on this structure due to the low homology between these sequences. Here, we describe the first crystal structures of hPLD1 and hPLD2 catalytic domains and identify novel structural elements and functional differences between the prokaryotic and eukaryotic enzymes. Furthermore, structure-based mutation studies and structures of inhibitor–hPLD complexes allowed us to elucidate the binding modes of dual and isoform-selective inhibitors, highlight key determinants of isoenzyme selectivity and provide a basis for further structure-based drug discovery and functional characterization of this therapeutically important superfamily of enzymes.
中文翻译:

人类 PLD 结构使药物设计和同工酶选择性表征成为可能
磷脂酶 D 酶 (PLD) 是普遍存在的磷酸二酯酶,可产生磷脂酸 (PA),这是一种关键的第二信使和生物合成构件。虽然是一种直系同源细菌链霉菌sp。菌株 PMF PLD 结构在 20 年前被解决,由于这些序列之间的低同源性,基于这种结构的人类 PLD 酶 (hPLD) 功能的分子基础仍然不清楚。在这里,我们描述了 hPLD1 和 hPLD2 催化结构域的第一个晶体结构,并确定了原核和真核酶之间的新结构元素和功能差异。此外,基于结构的突变研究和抑制剂-hPLD复合物的结构使我们能够阐明双重和异构体选择性抑制剂的结合模式,突出同工酶选择性的关键决定因素,并为进一步基于结构的药物发现和功能表征提供基础。这个具有治疗意义的酶超家族。



























 京公网安备 11010802027423号
京公网安备 11010802027423号