当前位置:
X-MOL 学术
›
Genes. Immun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Genomic profiling of intestinal T-cell receptor repertoires in inflammatory bowel disease.
Genes and Immunity ( IF 5 ) Pub Date : 2020-02-07 , DOI: 10.1038/s41435-020-0092-x Keerthana Saravanarajan 1 , Atiyekeogbebe Rita Douglas 2 , Mohd Syafiq Ismail 2 , Joseph Omorogbe 2 , Serhiy Semenov 2 , Greg Muphy 2 , Fiona O'Riordan 2 , Deirdre McNamara 2 , Shigeki Nakagome 1, 3
Genes and Immunity ( IF 5 ) Pub Date : 2020-02-07 , DOI: 10.1038/s41435-020-0092-x Keerthana Saravanarajan 1 , Atiyekeogbebe Rita Douglas 2 , Mohd Syafiq Ismail 2 , Joseph Omorogbe 2 , Serhiy Semenov 2 , Greg Muphy 2 , Fiona O'Riordan 2 , Deirdre McNamara 2 , Shigeki Nakagome 1, 3
Affiliation
|
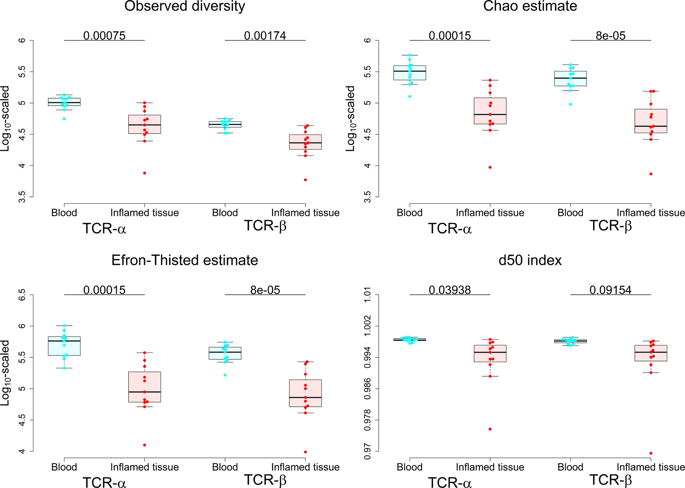
Growing evidence shows that inflammatory bowel disease (IBD) results from dysregulation of immune responses to gut microbes. T-cell receptors (TCRs) expressed on the T-cell surface play critical roles in discriminating pathogens from commensal intestinal microorganisms at the front line of the adaptive immune system. The breakdown of this interaction may trigger persistent inflammatory responses to gut bacteria, resulting in IBD. Taking advantage of high-throughput sequencing, we developed an integrated approach to dissect the intestinal TCR repertoires underlying IBD by collecting peripheral blood and inflamed intestine from the same set of 11 IBD cases. The intestinal TCR repertoires show lower clonotype diversity (p < 0.05) and stronger clonal expansion (p < 0.02) than those in the blood. This pattern becomes more profound in TCRs unique to the inflamed tissue compared with shared TCRs. Our approach further identified the increased usage of TRAV12-3 (false discovery rate, FDR < 5%), which biases its choices of J genes towards the reduction of TRAJ37 and TRAJ43 usage (FDR < 20%) in the inflamed intestine. Our genomic profiling suggests that this selective bias of V and J gene usage may lead to a loss of diversity in the intestinal TCR repertoires and result in mucosal inflammation in IBD.
中文翻译:

炎症性肠病中肠T细胞受体库的基因组分析。
越来越多的证据表明,炎症性肠病(IBD)是由肠道微生物免疫反应失调导致的。T细胞表面上表达的T细胞受体(TCR)在区分病原体与适应性免疫系统前线的共生肠道微生物中起关键作用。这种相互作用的破坏可能触发对肠道细菌的持续炎症反应,从而导致IBD。利用高通量测序的优势,我们开发了一种集成方法,可通过从11例IBD病例中收集外周血和发炎的肠道来解剖IBD的肠道TCR组成。与血液相比,肠道TCR谱表显示出更低的克隆型多样性(p <0.05)和更强的克隆扩增能力(p <0.02)。与共享的TCR相比,这种模式在发炎组织特有的TCR中变得更加深刻。我们的方法进一步确定了TRAV12-3的使用量增加(错误发现率,FDR <5%),这将其J基因的选择偏向于减少发炎肠中TRAJ37和TRAJ43的使用(FDR <20%)。我们的基因组分析表明,V和J基因使用的这种选择性偏倚可能会导致肠道TCR组成成分多样性的丧失,并导致IBD的粘膜炎症。
更新日期:2020-02-07
中文翻译:

炎症性肠病中肠T细胞受体库的基因组分析。
越来越多的证据表明,炎症性肠病(IBD)是由肠道微生物免疫反应失调导致的。T细胞表面上表达的T细胞受体(TCR)在区分病原体与适应性免疫系统前线的共生肠道微生物中起关键作用。这种相互作用的破坏可能触发对肠道细菌的持续炎症反应,从而导致IBD。利用高通量测序的优势,我们开发了一种集成方法,可通过从11例IBD病例中收集外周血和发炎的肠道来解剖IBD的肠道TCR组成。与血液相比,肠道TCR谱表显示出更低的克隆型多样性(p <0.05)和更强的克隆扩增能力(p <0.02)。与共享的TCR相比,这种模式在发炎组织特有的TCR中变得更加深刻。我们的方法进一步确定了TRAV12-3的使用量增加(错误发现率,FDR <5%),这将其J基因的选择偏向于减少发炎肠中TRAJ37和TRAJ43的使用(FDR <20%)。我们的基因组分析表明,V和J基因使用的这种选择性偏倚可能会导致肠道TCR组成成分多样性的丧失,并导致IBD的粘膜炎症。


























 京公网安备 11010802027423号
京公网安备 11010802027423号