当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
De Novo Designed Protein-Interaction Modules for In-Cell Applications.
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-02-06 , DOI: 10.1021/acssynbio.9b00453 Caitlin L Edgell 1, 2 , Abigail J Smith 2, 3 , Joseph L Beesley 1 , Nigel J Savery 2, 3 , Derek N Woolfson 1, 2, 3
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-02-06 , DOI: 10.1021/acssynbio.9b00453 Caitlin L Edgell 1, 2 , Abigail J Smith 2, 3 , Joseph L Beesley 1 , Nigel J Savery 2, 3 , Derek N Woolfson 1, 2, 3
Affiliation
|
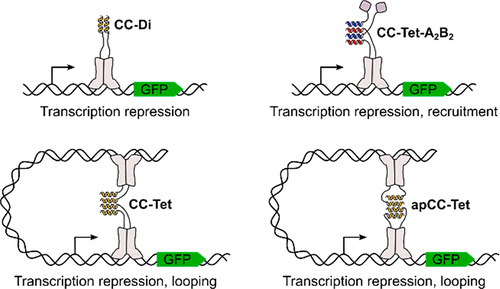
Protein-protein interactions control a wide variety of natural biological processes. α-Helical coiled coils frequently mediate such protein-protein interactions. Due to the relative simplicity of their sequences and structures and the ease with which properties such as strength and specificity of interaction can be controlled, coiled coils can be designed de novo to deliver a variety of non-natural protein-protein interaction domains. Herein, several de novo designed coiled coils are tested for their ability to mediate protein-protein interactions in Escherichia coli cells. The set includes a parallel homodimer, a parallel homotetramer, an antiparallel homotetramer, and a newly designed heterotetramer, all of which have been characterized in vitro by biophysical and structural methods. Using a transcription repression assay based on reconstituting the Lac repressor, we find that the modules behave as designed in the cellular environment. Each design imparts a different property to the resulting Lac repressor-coiled coil complexes, resulting in the benefit of being able to reconfigure the system in multiple ways. Modification of the system also allows the interactions to be controlled: assembly can be tuned by controlling the expression of the constituent components, and complexes can be disrupted through helix sequestration. The small and straightforward de novo designed components that we deliver are highly versatile and have considerable potential as protein-protein interaction domains in synthetic biology where proteins must be assembled in highly specific ways. The relative simplicity of the designs makes them amenable to future modifications to introduce finer control over their assembly and to adapt them for different contexts.
中文翻译:

从头设计用于细胞内应用的蛋白质相互作用模块。
蛋白质-蛋白质相互作用控制着各种各样的自然生物过程。α-螺旋螺旋经常介导这种蛋白质-蛋白质相互作用。由于它们的序列和结构相对简单,并且可以轻松控制相互作用的强度和特异性等特性,因此可以从头设计卷曲螺旋以提供各种非天然蛋白质-蛋白质相互作用结构域。在此,测试了几种从头设计的螺旋线圈在大肠杆菌细胞中介导蛋白质-蛋白质相互作用的能力。该集合包括一个平行同源二聚体、一个平行同源四聚体、一个反平行同源四聚体和一个新设计的异源四聚体,所有这些都已通过生物物理和结构方法进行了体外表征。使用基于重组 Lac 阻遏物的转录抑制测定,我们发现模块在细胞环境中的行为与设计的一样。每种设计都赋予所产生的 Lac 阻遏物螺旋线圈复合物不同的特性,从而带来能够以多种方式重新配置系统的好处。系统的修改还允许控制相互作用:可以通过控制组成成分的表达来调整组装,并且可以通过螺旋隔离来破坏复合物。我们提供的小而直接的从头设计组件具有高度通用性,并且在合成生物学中作为蛋白质-蛋白质相互作用域具有相当大的潜力,其中蛋白质必须以高度特定的方式组装。
更新日期:2020-02-06
中文翻译:

从头设计用于细胞内应用的蛋白质相互作用模块。
蛋白质-蛋白质相互作用控制着各种各样的自然生物过程。α-螺旋螺旋经常介导这种蛋白质-蛋白质相互作用。由于它们的序列和结构相对简单,并且可以轻松控制相互作用的强度和特异性等特性,因此可以从头设计卷曲螺旋以提供各种非天然蛋白质-蛋白质相互作用结构域。在此,测试了几种从头设计的螺旋线圈在大肠杆菌细胞中介导蛋白质-蛋白质相互作用的能力。该集合包括一个平行同源二聚体、一个平行同源四聚体、一个反平行同源四聚体和一个新设计的异源四聚体,所有这些都已通过生物物理和结构方法进行了体外表征。使用基于重组 Lac 阻遏物的转录抑制测定,我们发现模块在细胞环境中的行为与设计的一样。每种设计都赋予所产生的 Lac 阻遏物螺旋线圈复合物不同的特性,从而带来能够以多种方式重新配置系统的好处。系统的修改还允许控制相互作用:可以通过控制组成成分的表达来调整组装,并且可以通过螺旋隔离来破坏复合物。我们提供的小而直接的从头设计组件具有高度通用性,并且在合成生物学中作为蛋白质-蛋白质相互作用域具有相当大的潜力,其中蛋白质必须以高度特定的方式组装。



























 京公网安备 11010802027423号
京公网安备 11010802027423号