Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Dysregulation of multiple metabolic networks related to brain transmethylation and polyamine pathways in Alzheimer disease: A targeted metabolomic and transcriptomic study.
PLOS Medicine ( IF 15.8 ) Pub Date : 2020-01-24 , DOI: 10.1371/journal.pmed.1003012 Uma V Mahajan 1 , Vijay R Varma 1 , Michael E Griswold 2 , Chad T Blackshear 2 , Yang An 3 , Anup M Oommen 4 , Sudhir Varma 5 , Juan C Troncoso 6 , Olga Pletnikova 6 , Richard O'Brien 7 , Timothy J Hohman 8 , Cristina Legido-Quigley 9, 10 , Madhav Thambisetty 1
PLOS Medicine ( IF 15.8 ) Pub Date : 2020-01-24 , DOI: 10.1371/journal.pmed.1003012 Uma V Mahajan 1 , Vijay R Varma 1 , Michael E Griswold 2 , Chad T Blackshear 2 , Yang An 3 , Anup M Oommen 4 , Sudhir Varma 5 , Juan C Troncoso 6 , Olga Pletnikova 6 , Richard O'Brien 7 , Timothy J Hohman 8 , Cristina Legido-Quigley 9, 10 , Madhav Thambisetty 1
Affiliation
|
BACKGROUND
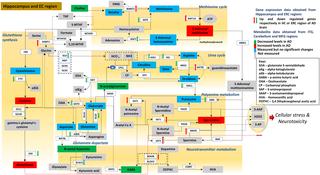
There is growing evidence that Alzheimer disease (AD) is a pervasive metabolic disorder with dysregulation in multiple biochemical pathways underlying its pathogenesis. Understanding how perturbations in metabolism are related to AD is critical to identifying novel targets for disease-modifying therapies. In this study, we test whether AD pathogenesis is associated with dysregulation in brain transmethylation and polyamine pathways.
METHODS AND FINDINGS
We first performed targeted and quantitative metabolomics assays using capillary electrophoresis-mass spectrometry (CE-MS) on brain samples from three groups in the Baltimore Longitudinal Study of Aging (BLSA) (AD: n = 17; Asymptomatic AD [ASY]: n = 13; Control [CN]: n = 13) (overall 37.2% female; mean age at death 86.118 ± 9.842 years) in regions both vulnerable and resistant to AD pathology. Using linear mixed-effects models within two primary brain regions (inferior temporal gyrus [ITG] and middle frontal gyrus [MFG]), we tested associations between brain tissue concentrations of 26 metabolites and the following primary outcomes: group differences, Consortium to Establish a Registry for Alzheimer's Disease (CERAD) (neuritic plaque burden), and Braak (neurofibrillary pathology) scores. We found significant alterations in concentrations of metabolites in AD relative to CN samples, as well as associations with severity of both CERAD and Braak, mainly in the ITG. These metabolites represented biochemical reactions in the (1) methionine cycle (choline: lower in AD, p = 0.003; S-adenosyl methionine: higher in AD, p = 0.005); (2) transsulfuration and glutathione synthesis (cysteine: higher in AD, p < 0.001; reduced glutathione [GSH]: higher in AD, p < 0.001); (3) polyamine synthesis/catabolism (spermidine: higher in AD, p = 0.004); (4) urea cycle (N-acetyl glutamate: lower in AD, p < 0.001); (5) glutamate-aspartate metabolism (N-acetyl aspartate: lower in AD, p = 0.002); and (6) neurotransmitter metabolism (gamma-amino-butyric acid: lower in AD, p < 0.001). Utilizing three Gene Expression Omnibus (GEO) datasets, we then examined mRNA expression levels of 71 genes encoding enzymes regulating key reactions within these pathways in the entorhinal cortex (ERC; AD: n = 25; CN: n = 52) and hippocampus (AD: n = 29; CN: n = 56). Complementing our metabolomics results, our transcriptomics analyses also revealed significant alterations in gene expression levels of key enzymatic regulators of biochemical reactions linked to transmethylation and polyamine metabolism. Our study has limitations: our metabolomics assays measured only a small proportion of all metabolites participating in the pathways we examined. Our study is also cross-sectional, limiting our ability to directly test how AD progression may impact changes in metabolite concentrations or differential-gene expression. Additionally, the relatively small number of brain tissue samples may have limited our power to detect alterations in all pathway-specific metabolites and their genetic regulators.
CONCLUSIONS
In this study, we observed broad dysregulation of transmethylation and polyamine synthesis/catabolism, including abnormalities in neurotransmitter signaling, urea cycle, aspartate-glutamate metabolism, and glutathione synthesis. Our results implicate alterations in cellular methylation potential and increased flux in the transmethylation pathways, increased demand on antioxidant defense mechanisms, perturbations in intermediate metabolism in the urea cycle and aspartate-glutamate pathways disrupting mitochondrial bioenergetics, increased polyamine biosynthesis and breakdown, as well as abnormalities in neurotransmitter metabolism that are related to AD.
中文翻译:

与阿尔茨海默病脑转甲基化和多胺途径相关的多个代谢网络失调:一项针对性的代谢组学和转录组学研究。
背景技术越来越多的证据表明,阿尔茨海默氏病(AD)是一种普遍的代谢紊乱,在其发病机理的多个生化途径中存在异常调节。了解新陈代谢的扰动与AD的关系,对于确定疾病改良疗法的新靶标至关重要。在这项研究中,我们测试了AD的发病机理是否与大脑甲基化和多胺途径的失调有关。方法和研究结果我们首先在巴尔的摩纵向老龄化研究(BLSA)中对三组脑样本进行了毛细管电泳-质谱(CE-MS)靶向和定量代谢组学测定(AD:n = 17;无症状AD [ASY] :n = 13;对照组[CN]:n = 13)(女性占总人数的37.2%;平均死亡年龄为86.118±9。842岁)在既易受AD病理影响又对AD病理具有抵抗力的地区。使用两个主要大脑区域(下颞回[ITG]和额中回[MFG])内的线性混合效应模型,我们测试了26种代谢产物的脑组织浓度与以下主要结果之间的关联:组差异,建立联合体的财团阿尔茨海默氏病(CERAD)(神经斑块负担)和Braak(神经原纤维病理学)评分的注册表。我们发现相对于CN样品,AD中代谢物浓度的显着变化,以及与CERAD和Braak严重程度的相关性(主要在ITG中)。这些代谢物代表了(1)蛋氨酸循环中的生化反应(胆碱:AD较低,p = 0.003; S-腺苷甲硫氨酸:AD较高,p = 0.005);(2)转硫和谷胱甘肽合成(半胱氨酸:AD较高,p <0.001;还原型谷胱甘肽[GSH]:AD较高,p <0.001);(3)多胺的合成/分解代谢(亚精胺:AD较高,p = 0.004);(4)尿素循环(N-乙酰谷氨酸:AD较低,p <0.001);(5)谷氨酸-天冬氨酸代谢(N-乙酰天冬氨酸:AD较低,p = 0.002);(6)神经递质代谢(γ-氨基丁酸:AD较低,p <0.001)。利用三个基因表达综合(GEO)数据集,我们随后检查了71种基因的mRNA表达水平,这些基因编码调节内嗅皮层(ERC; AD:n = 25; CN:n = 52)和海马(AD)这些途径内关键反应的酶。 :n = 29; CN:n = 56)。补充我们的代谢组学结果,我们的转录组学分析还发现,与转甲基化和多胺代谢有关的生化反应的关键酶调节剂的基因表达水平发生了显着变化。我们的研究存在局限性:我们的代谢组学测定仅测量了参与我们所研究途径的所有代谢物的一小部分。我们的研究也是横断面的,限制了我们直接测试AD进展如何影响代谢物浓度或差异基因表达变化的能力。此外,相对较少的脑组织样本可能限制了我们检测所有途径特异性代谢物及其遗传调节剂变化的能力。结论在这项研究中,我们观察到甲基化和聚胺合成/分解代谢的广泛失调,包括神经递质信号传导,尿素循环,天冬氨酸-谷氨酸代谢和谷胱甘肽合成异常。我们的研究结果暗示了细胞甲基化潜力的改变和甲基转移途径中通量的增加,对抗氧化剂防御机制的需求增加,尿素循环中中间代谢的扰动以及天冬氨酸-谷氨酸途径破坏线粒体生物能学,多胺生物合成和分解增加以及异常与AD有关的神经递质代谢
更新日期:2020-01-26
中文翻译:

与阿尔茨海默病脑转甲基化和多胺途径相关的多个代谢网络失调:一项针对性的代谢组学和转录组学研究。
背景技术越来越多的证据表明,阿尔茨海默氏病(AD)是一种普遍的代谢紊乱,在其发病机理的多个生化途径中存在异常调节。了解新陈代谢的扰动与AD的关系,对于确定疾病改良疗法的新靶标至关重要。在这项研究中,我们测试了AD的发病机理是否与大脑甲基化和多胺途径的失调有关。方法和研究结果我们首先在巴尔的摩纵向老龄化研究(BLSA)中对三组脑样本进行了毛细管电泳-质谱(CE-MS)靶向和定量代谢组学测定(AD:n = 17;无症状AD [ASY] :n = 13;对照组[CN]:n = 13)(女性占总人数的37.2%;平均死亡年龄为86.118±9。842岁)在既易受AD病理影响又对AD病理具有抵抗力的地区。使用两个主要大脑区域(下颞回[ITG]和额中回[MFG])内的线性混合效应模型,我们测试了26种代谢产物的脑组织浓度与以下主要结果之间的关联:组差异,建立联合体的财团阿尔茨海默氏病(CERAD)(神经斑块负担)和Braak(神经原纤维病理学)评分的注册表。我们发现相对于CN样品,AD中代谢物浓度的显着变化,以及与CERAD和Braak严重程度的相关性(主要在ITG中)。这些代谢物代表了(1)蛋氨酸循环中的生化反应(胆碱:AD较低,p = 0.003; S-腺苷甲硫氨酸:AD较高,p = 0.005);(2)转硫和谷胱甘肽合成(半胱氨酸:AD较高,p <0.001;还原型谷胱甘肽[GSH]:AD较高,p <0.001);(3)多胺的合成/分解代谢(亚精胺:AD较高,p = 0.004);(4)尿素循环(N-乙酰谷氨酸:AD较低,p <0.001);(5)谷氨酸-天冬氨酸代谢(N-乙酰天冬氨酸:AD较低,p = 0.002);(6)神经递质代谢(γ-氨基丁酸:AD较低,p <0.001)。利用三个基因表达综合(GEO)数据集,我们随后检查了71种基因的mRNA表达水平,这些基因编码调节内嗅皮层(ERC; AD:n = 25; CN:n = 52)和海马(AD)这些途径内关键反应的酶。 :n = 29; CN:n = 56)。补充我们的代谢组学结果,我们的转录组学分析还发现,与转甲基化和多胺代谢有关的生化反应的关键酶调节剂的基因表达水平发生了显着变化。我们的研究存在局限性:我们的代谢组学测定仅测量了参与我们所研究途径的所有代谢物的一小部分。我们的研究也是横断面的,限制了我们直接测试AD进展如何影响代谢物浓度或差异基因表达变化的能力。此外,相对较少的脑组织样本可能限制了我们检测所有途径特异性代谢物及其遗传调节剂变化的能力。结论在这项研究中,我们观察到甲基化和聚胺合成/分解代谢的广泛失调,包括神经递质信号传导,尿素循环,天冬氨酸-谷氨酸代谢和谷胱甘肽合成异常。我们的研究结果暗示了细胞甲基化潜力的改变和甲基转移途径中通量的增加,对抗氧化剂防御机制的需求增加,尿素循环中中间代谢的扰动以及天冬氨酸-谷氨酸途径破坏线粒体生物能学,多胺生物合成和分解增加以及异常与AD有关的神经递质代谢



























 京公网安备 11010802027423号
京公网安备 11010802027423号