Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
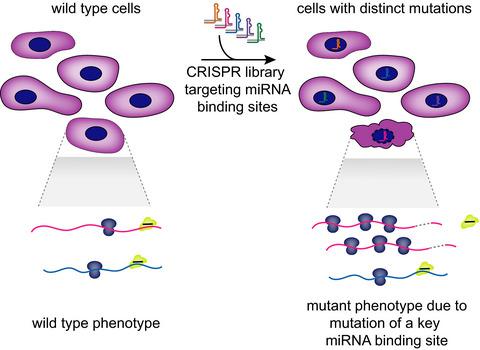
CRISPR screening strategies for microRNA target identification.
The FEBS Journal ( IF 5.4 ) Pub Date : 2020-01-23 , DOI: 10.1111/febs.15218 Bing Yang 1 , Katherine McJunkin 1
The FEBS Journal ( IF 5.4 ) Pub Date : 2020-01-23 , DOI: 10.1111/febs.15218 Bing Yang 1 , Katherine McJunkin 1
Affiliation
|
Identifying microRNA (miRNA) target genes remains a major challenge in understanding the roles miRNAs play in gene regulation. Furthermore, understanding which miRNA–target interactions are the most biologically important is even more difficult. We present CRISPR‐based strategies to identify essential miRNA binding sites. First, CRISPR knockout screens can easily be adapted to identify genes whose inactivation suppresses miRNA mutant phenotypes. Second, a custom approach to target individual miRNA binding sites via CRISPR can identify sites whose mutation recapitulates miRNA mutant phenotypes. We emphasize that the latter approach requires a readout of mutational profile (rather than single guide RNA abundance) when applied in a negative selection setting. Overall, the advent of CRISPR technology alongside improving empirical means of miRNA target identification will accelerate our dissection of miRNA gene regulatory networks.
中文翻译:

用于microRNA目标识别的CRISPR筛选策略。
在了解miRNA在基因调控中的作用时,鉴定microRNA(miRNA)靶基因仍然是一项重大挑战。此外,要了解哪种miRNA与靶标之间的相互作用在生物学上最重要,就更加困难。我们提出了基于CRISPR的策略来识别必要的miRNA结合位点。首先,CRISPR基因敲除筛选可以轻松地用于鉴定其失活抑制miRNA突变表型的基因。其次,通过CRISPR靶向单个miRNA结合位点的定制方法可以识别其突变概括了miRNA突变表型的位点。我们强调,后一种方法在阴性选择条件下应用时,需要读出突变谱(而不是单个向导RNA的丰度)。总体,
更新日期:2020-01-23
中文翻译:

用于microRNA目标识别的CRISPR筛选策略。
在了解miRNA在基因调控中的作用时,鉴定microRNA(miRNA)靶基因仍然是一项重大挑战。此外,要了解哪种miRNA与靶标之间的相互作用在生物学上最重要,就更加困难。我们提出了基于CRISPR的策略来识别必要的miRNA结合位点。首先,CRISPR基因敲除筛选可以轻松地用于鉴定其失活抑制miRNA突变表型的基因。其次,通过CRISPR靶向单个miRNA结合位点的定制方法可以识别其突变概括了miRNA突变表型的位点。我们强调,后一种方法在阴性选择条件下应用时,需要读出突变谱(而不是单个向导RNA的丰度)。总体,



























 京公网安备 11010802027423号
京公网安备 11010802027423号