Nature Chemistry ( IF 21.8 ) Pub Date : 2020-01-20 , DOI: 10.1038/s41557-019-0406-7 Di Liu 1 , Cody W Geary 2, 3 , Gang Chen 1, 4 , Yaming Shao 5 , Mo Li 6 , Chengde Mao 6 , Ebbe S Andersen 2 , Joseph A Piccirilli 1, 5 , Paul W K Rothemund 3 , Yossi Weizmann 1, 7
|
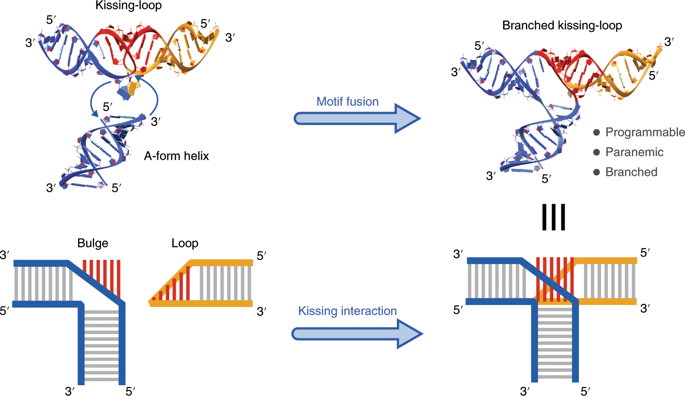
In biological systems, large and complex structures are often assembled from multiple simpler identical subunits. This strategy—homooligomerization—allows efficient genetic encoding of structures and avoids the need to control the stoichiometry of multiple distinct units. It also allows the minimal number of distinct subunits when designing artificial nucleic acid structures. Here, we present a robust self-assembly system in which homooligomerizable tiles are formed from intramolecularly folded RNA single strands. Tiles are linked through an artificially designed branched kissing-loop motif, involving Watson–Crick base pairing between the single-stranded regions of a bulged helix and a hairpin loop. By adjusting the tile geometry to gain control over the curvature, torsion and the number of helices, we have constructed 16 different linear and circular structures, including a finite-sized three-dimensional cage. We further demonstrate cotranscriptional self-assembly of tiles based on branched kissing loops, and show that tiles inserted into a transfer RNA scaffold can be overexpressed in bacterial cells.
中文翻译:

用于构建多种 RNA 同源寡聚纳米结构的分支接吻环
在生物系统中,大而复杂的结构通常由多个更简单的相同亚基组装而成。这种策略——同寡聚化——允许对结构进行有效的遗传编码,并避免需要控制多个不同单元的化学计量。在设计人工核酸结构时,它还允许最少数量的不同亚基。在这里,我们提出了一个强大的自组装系统,其中同源寡聚瓷砖由分子内折叠的 RNA 单链形成。瓷砖通过人工设计的分支接吻环基序连接,包括凸出螺旋和发夹环的单链区域之间的 Watson-Crick 碱基配对。通过调整瓷砖几何形状来控制曲率、扭转和螺旋的数量,我们构建了 16 种不同的线性和圆形结构,包括一个有限尺寸的三维笼子。我们进一步证明了基于分支接吻环的瓷砖的共转录自组装,并表明插入转移 RNA 支架的瓷砖可以在细菌细胞中过度表达。


























 京公网安备 11010802027423号
京公网安备 11010802027423号