当前位置:
X-MOL 学术
›
J. Biomed. Inform.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Differential splicing analysis based on isoforms expression with NBSplice.
Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2020-01-21 , DOI: 10.1016/j.jbi.2020.103378 Gabriela Alejandra Merino 1 , Elmer Andrés Fernández 2
Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2020-01-21 , DOI: 10.1016/j.jbi.2020.103378 Gabriela Alejandra Merino 1 , Elmer Andrés Fernández 2
Affiliation
|
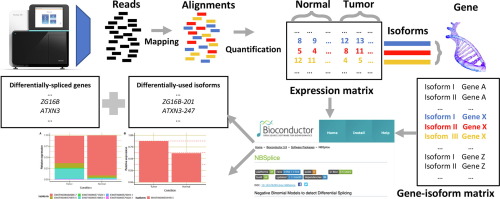
Alternative splicing alterations have been widely related to several human diseases revealing the importance of their study for the success of translational medicine. Differential splicing (DS) occurrence has been mainly analyzed through exon-based approaches over RNA-seq data. Although these strategies allow identifying differentially spliced genes, they ignore the identity of the affected gene isoforms which is crucial to understand the underlying pathological processes behind alternative splicing changes. Moreover, despite several isoform quantification tools for RNA-seq data have been recently developed, DS tools have not taken advantage of them. Here, the NBSplice R package for differential splicing analysis by means of isoform expression data is presented. It estimates differences on relative expressions of gene transcripts between experimental conditions to infer changes in gene alternative splicing patterns. The developed tool was evaluated using a synthetic RNA-seq dataset with controlled differential splicing. NBSplice accurately predicted DS occurrence, outperforming current methods in terms of accuracy, sensitivity, F-score, and false discovery rate control. The usefulness of our development was demonstrated by the analysis of a real cancer dataset, revealing new differentially spliced genes that could be studied pursuing new colorectal cancer biomarkers discovery.
中文翻译:

基于NBSplice的同工型表达的差异剪接分析。
选择性剪接改变已与几种人类疾病广泛相关,从而揭示了其研究对转化医学成功的重要性。主要通过基于外显子的方法对RNA-seq数据分析了差异剪接(DS)的发生。尽管这些策略允许鉴定差异剪接的基因,但它们忽略了受影响的基因同工型的身份,这对于理解替代剪接变化背后的潜在病理过程至关重要。而且,尽管最近已经开发了几种用于RNA-seq数据的同工型定量工具,但DS工具并未利用它们。在这里,介绍了NBSplice R软件包,用于通过同工型表达数据进行差异剪接分析。它估计了实验条件之间基因转录本相对表达的差异,以推断基因替代剪接模式的变化。使用具有受控差异剪接的合成RNA-seq数据集评估了开发的工具。NBSplice准确地预测了DS的发生,在准确性,灵敏度,F得分和错误发现率控制方面均优于当前方法。通过对真实癌症数据集的分析证明了我们开发的有用性,揭示了新的差异剪接基因,可以通过研究发现新的结直肠癌生物标记物。在准确性,灵敏度,F分数和错误发现率控制方面均胜过当前方法。通过对真实癌症数据集的分析证明了我们开发的有用性,揭示了新的差异剪接基因,可以通过研究发现新的结直肠癌生物标记物。在准确性,灵敏度,F分数和错误发现率控制方面均胜过当前方法。通过对真实癌症数据集的分析证明了我们开发的有用性,揭示了新的差异剪接基因,可以通过研究发现新的结直肠癌生物标记物。
更新日期:2020-01-21
中文翻译:

基于NBSplice的同工型表达的差异剪接分析。
选择性剪接改变已与几种人类疾病广泛相关,从而揭示了其研究对转化医学成功的重要性。主要通过基于外显子的方法对RNA-seq数据分析了差异剪接(DS)的发生。尽管这些策略允许鉴定差异剪接的基因,但它们忽略了受影响的基因同工型的身份,这对于理解替代剪接变化背后的潜在病理过程至关重要。而且,尽管最近已经开发了几种用于RNA-seq数据的同工型定量工具,但DS工具并未利用它们。在这里,介绍了NBSplice R软件包,用于通过同工型表达数据进行差异剪接分析。它估计了实验条件之间基因转录本相对表达的差异,以推断基因替代剪接模式的变化。使用具有受控差异剪接的合成RNA-seq数据集评估了开发的工具。NBSplice准确地预测了DS的发生,在准确性,灵敏度,F得分和错误发现率控制方面均优于当前方法。通过对真实癌症数据集的分析证明了我们开发的有用性,揭示了新的差异剪接基因,可以通过研究发现新的结直肠癌生物标记物。在准确性,灵敏度,F分数和错误发现率控制方面均胜过当前方法。通过对真实癌症数据集的分析证明了我们开发的有用性,揭示了新的差异剪接基因,可以通过研究发现新的结直肠癌生物标记物。在准确性,灵敏度,F分数和错误发现率控制方面均胜过当前方法。通过对真实癌症数据集的分析证明了我们开发的有用性,揭示了新的差异剪接基因,可以通过研究发现新的结直肠癌生物标记物。


























 京公网安备 11010802027423号
京公网安备 11010802027423号