当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Generating quantitative binding landscapes through fractional binding selections combined with deep sequencing and data normalization.
Nature Communications ( IF 16.6 ) Pub Date : 2020-01-15 , DOI: 10.1038/s41467-019-13895-8 Michael Heyne 1, 2 , Niv Papo 2 , Julia M Shifman 1
Nature Communications ( IF 16.6 ) Pub Date : 2020-01-15 , DOI: 10.1038/s41467-019-13895-8 Michael Heyne 1, 2 , Niv Papo 2 , Julia M Shifman 1
Affiliation
|
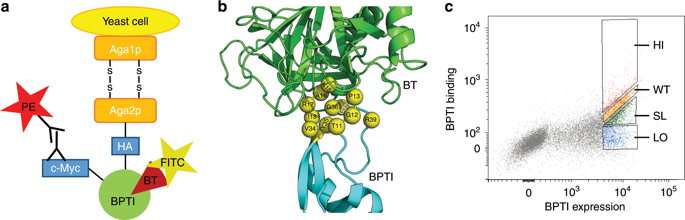
Quantifying the effects of various mutations on binding free energy is crucial for understanding the evolution of protein-protein interactions and would greatly facilitate protein engineering studies. Yet, measuring changes in binding free energy (ΔΔGbind) remains a tedious task that requires expression of each mutant, its purification, and affinity measurements. We developed an attractive approach that allows us to quantify ΔΔGbind for thousands of protein mutants in one experiment. Our protocol combines protein randomization, Yeast Surface Display technology, deep sequencing, and a few experimental ΔΔGbind data points on purified proteins to generate ΔΔGbind values for the remaining numerous mutants of the same protein complex. Using this methodology, we comprehensively map the single-mutant binding landscape of one of the highest-affinity interaction between BPTI and Bovine Trypsin (BT). We show that ΔΔGbind for this interaction could be quantified with high accuracy over the range of 12 kcal mol-1 displayed by various BPTI single mutants.
中文翻译:

通过分数结合选择结合深度测序和数据归一化生成定量结合图。
量化各种突变对结合自由能的影响对于理解蛋白质-蛋白质相互作用的演化至关重要,并且将极大地促进蛋白质工程研究。然而,测量结合自由能(ΔΔGbind)的变化仍然是繁琐的工作,需要每个突变体的表达,其纯化和亲和力测量。我们开发了一种有吸引力的方法,使我们可以在一个实验中对数千种蛋白质突变体进行ΔΔGbind量化。我们的协议结合了蛋白质随机化,酵母表面展示技术,深度测序和纯化蛋白质上的一些实验性ΔΔGbind数据点,以为同一蛋白质复合物的其余众多突变体生成ΔΔGbind值。使用这种方法,我们全面绘制了BPTI和牛胰蛋白酶(BT)之间最高亲和力相互作用之一的单突变结合图。我们表明,可以在各种BPTI单突变体显示的12 kcal mol-1范围内以高精度对这种相互作用的ΔΔGbind进行定量。
更新日期:2020-01-15
中文翻译:

通过分数结合选择结合深度测序和数据归一化生成定量结合图。
量化各种突变对结合自由能的影响对于理解蛋白质-蛋白质相互作用的演化至关重要,并且将极大地促进蛋白质工程研究。然而,测量结合自由能(ΔΔGbind)的变化仍然是繁琐的工作,需要每个突变体的表达,其纯化和亲和力测量。我们开发了一种有吸引力的方法,使我们可以在一个实验中对数千种蛋白质突变体进行ΔΔGbind量化。我们的协议结合了蛋白质随机化,酵母表面展示技术,深度测序和纯化蛋白质上的一些实验性ΔΔGbind数据点,以为同一蛋白质复合物的其余众多突变体生成ΔΔGbind值。使用这种方法,我们全面绘制了BPTI和牛胰蛋白酶(BT)之间最高亲和力相互作用之一的单突变结合图。我们表明,可以在各种BPTI单突变体显示的12 kcal mol-1范围内以高精度对这种相互作用的ΔΔGbind进行定量。


























 京公网安备 11010802027423号
京公网安备 11010802027423号