Nature Protocols ( IF 14.8 ) Pub Date : 2020-01-15 , DOI: 10.1038/s41596-019-0250-7 Daphne S Bindels 1 , Marten Postma 1 , Lindsay Haarbosch 1 , Laura van Weeren , Theodorus W J Gadella 1
|
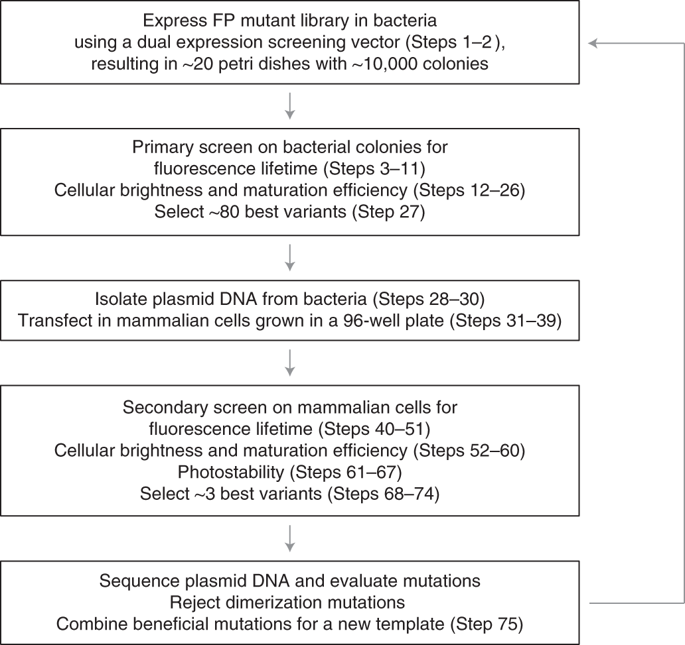
Genetically encoded fluorescent proteins (FPs) are highly utilized in cell biology research to study proteins of interest or signal processes using biosensors. To perform well in specific applications, these FPs require a multitude of tailored properties. It is for this reason that they need to be optimized by using mutagenesis. The optimization process through screening is often based solely on bacterial colony brightness, but multiple parameters ultimately determine the performance of an optimal FP. Instead of characterizing other properties after selection, we developed a multiparameter screening method based on four critical parametersscreened simultaneously: fluorescence lifetime, cellular brightness, maturation efficiency, and photostability. First, a high-throughput primary screen (based on fluorescence lifetime and cellular brightness using a mutated FP library) is performed in bacterial colonies. A secondary multiparameter screen based on all four parameters, using a novel bacterial–mammalian dual-expression vector enables expression of the best FP variants in mammalian cell lines. A newly developed automated multiparameter acquisition and cell-based analysis approach for 96-well plates further increased workflow efficiency. We used this protocol to yield the record-bright mScarlet, a fast-maturating mScarlet-I, and a photostable mScarlet-H. This protocol can also be applied to other FP classes or Förster resonance energy transfer (FRET)-based biosensors with minor adaptations. With an available mutant library of a template FP and a complete and tested laboratory setup, a single round of multiparameter screening (including the primary bacterial screen, secondary mammalian cell screen, sequencing, and data processing) can be performed within 2 weeks.
中文翻译:

用于开发优化的红色荧光蛋白的多参数筛选方法
基因编码的荧光蛋白 (FP) 在细胞生物学研究中被高度利用,以使用生物传感器研究感兴趣的蛋白质或信号过程。为了在特定应用中表现良好,这些 FP 需要大量定制的属性。正是出于这个原因,它们需要通过使用诱变进行优化。通过筛选的优化过程通常仅基于细菌菌落亮度,但多个参数最终决定了最佳 FP 的性能。我们没有在选择后表征其他特性,而是开发了一种基于同时筛选的四个关键参数的多参数筛选方法:荧光寿命、细胞亮度、成熟效率和光稳定性。第一的,在细菌菌落中进行高通量初级筛选(基于荧光寿命和使用突变 FP 库的细胞亮度)。基于所有四个参数的二级多参数筛选,使用新型细菌-哺乳动物双表达载体,能够在哺乳动物细胞系中表达最佳 FP 变体。新开发的用于 96 孔板的自动多参数采集和基于细胞的分析方法进一步提高了工作流程效率。我们使用该协议产生了创纪录的明亮 mScarlet、快速成熟的 mScarlet-I 和光稳定的 mScarlet-H。该协议还可以应用于其他 FP 类或基于 Förster 共振能量转移 (FRET) 的生物传感器,并进行轻微调整。使用模板 FP 的可用突变库和完整且经过测试的实验室设置,



























 京公网安备 11010802027423号
京公网安备 11010802027423号