Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
In silico analysis and high-risk pathogenic phenotype predictions of non-synonymous single nucleotide polymorphisms in human Crystallin beta A4 gene associated with congenital cataract.
PLOS ONE ( IF 3.7 ) Pub Date : 2020-01-14 , DOI: 10.1371/journal.pone.0227859 Zhenyu Wang 1, 2 , Chen Huang 1, 2, 3 , Huibin Lv 1, 2 , Mingzhou Zhang 1, 2 , Xuemin Li 1, 2
PLOS ONE ( IF 3.7 ) Pub Date : 2020-01-14 , DOI: 10.1371/journal.pone.0227859 Zhenyu Wang 1, 2 , Chen Huang 1, 2, 3 , Huibin Lv 1, 2 , Mingzhou Zhang 1, 2 , Xuemin Li 1, 2
Affiliation
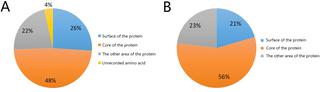
|
In order to provide a cost-effective method to narrow down the number of pathogenic Crystallin beta A4 (CRYBA4) non-synonymous single nucleotide polymorphisms (nsSNPs), we collected nsSNP information of the CRYBA4 gene from SNP databases and literature, predicting the pathogenicity and possible changes of protein properties and structures using multiple bioinformatics tools. The nsSNP data of the CRYBA4 gene were collected from 4 databases and published literature. According to 12 criteria, six bioinformatics tools were chosen to predict the pathogenicity. I-Mutant 2.0, Mupro and INPS online tools were used to analyze the effects of amino acid substitution on protein stability by calculating the value of ΔΔG. ConSurf, SOPMA, GETAREA and HOPE online tools were used to predict the evolutionary conservation of amino acids, solvent accessible surface areas, and the physical and chemical properties and changes of protein structure. All 157 CRYBA4 nsSNPs were analyzed. Forty-four CRYBA4 high-risk pathogenic nsSNPs (predicted to be pathogenic by all six software tools) were detected out of the 157 CRYBA4 nsSNPs, four of which (c.283C>T, p.R95W; c.449T>A, p.V150D; c.475G>A, p.G159R; c.575G>C, p.R192P) should be focused on because of their high potential pathogenicity and possibility of changing protein properties. Thirty high-risk nsSNPs were predicted to cause a decrease of protein stability. Twenty-nine high-risk nsSNPs occurred in evolutionary conserved positions. Twenty-two high-risk nsSNPs occurred in the core of the protein. It is predicted that these high-risk pathogenic nsSNPs can cause changes in the physical and chemical properties of amino acids, resulting in structural changes of proteins and changes in the interactions between domains and other molecules, thus affecting the function of proteins. This study provides important reference value when narrowing down the number of pathogenic CRYBA4 nsSNPs and studying the pathogenesis of congenital cataracts. By using this method, we can easily find 44 high-risk pathogenic nsSNPs out of 157 CRYBA4 nsSNPs.
中文翻译:

在计算机分析和高危致病性表型预测中与先天性白内障相关的人类Crystallin beta A4基因中非同义单核苷酸多态性。
为了提供一种经济有效的方法来缩小致病性Crystallin beta A4(CRYBA4)非同义单核苷酸多态性(nsSNPs)的数量,我们从SNP数据库和文献中收集了CRYBA4基因的nsSNP信息,预测了致病性和使用多种生物信息学工具可能会改变蛋白质特性和结构。从4个数据库中收集了CRYBA4基因的nsSNP数据,并发表了文献。根据12条标准,选择了6种生物信息学工具来预测致病性。使用I-Mutant 2.0,Mupro和INPS在线工具通过计算ΔΔG的值来分析氨基酸取代对蛋白质稳定性的影响。ConSurf,SOPMA,GETAREA和HOPE在线工具用于预测氨基酸的进化保守性,溶剂可及的表面积,以及理化性质和蛋白质结构的变化。分析了全部157个CRYBA4 nsSNP。在157个CRYBA4 nsSNP中检测出44个CRYBA4高风险致病性nsSNP(被全部六个软件工具预测为致病的),其中四个(c.283C> T,p.R95W; c.449T> A,p .V150D; c.475G> A,p.G159R; c.575G> C,p.R192P)应重点关注,因为它们具有很高的潜在致病性和改变蛋白质特性的可能性。预计有三十种高风险的nsSNP会导致蛋白质稳定性下降。二十九个高风险nsSNPs发生在进化保守的位置。该蛋白的核心中有22个高风险nsSNP。预计这些高风险的致病性nsSNP会导致氨基酸的物理和化学性质发生变化,导致蛋白质的结构变化以及结构域与其他分子之间相互作用的变化,从而影响蛋白质的功能。该研究为缩小致病性CRYBA4 nsSNPs的数量和研究先天性白内障的发病机理提供了重要的参考价值。通过这种方法,我们可以轻松地从157个CRYBA4 nsSNP中找到44个高致病性nsSNP。
更新日期:2020-01-15
中文翻译:

在计算机分析和高危致病性表型预测中与先天性白内障相关的人类Crystallin beta A4基因中非同义单核苷酸多态性。
为了提供一种经济有效的方法来缩小致病性Crystallin beta A4(CRYBA4)非同义单核苷酸多态性(nsSNPs)的数量,我们从SNP数据库和文献中收集了CRYBA4基因的nsSNP信息,预测了致病性和使用多种生物信息学工具可能会改变蛋白质特性和结构。从4个数据库中收集了CRYBA4基因的nsSNP数据,并发表了文献。根据12条标准,选择了6种生物信息学工具来预测致病性。使用I-Mutant 2.0,Mupro和INPS在线工具通过计算ΔΔG的值来分析氨基酸取代对蛋白质稳定性的影响。ConSurf,SOPMA,GETAREA和HOPE在线工具用于预测氨基酸的进化保守性,溶剂可及的表面积,以及理化性质和蛋白质结构的变化。分析了全部157个CRYBA4 nsSNP。在157个CRYBA4 nsSNP中检测出44个CRYBA4高风险致病性nsSNP(被全部六个软件工具预测为致病的),其中四个(c.283C> T,p.R95W; c.449T> A,p .V150D; c.475G> A,p.G159R; c.575G> C,p.R192P)应重点关注,因为它们具有很高的潜在致病性和改变蛋白质特性的可能性。预计有三十种高风险的nsSNP会导致蛋白质稳定性下降。二十九个高风险nsSNPs发生在进化保守的位置。该蛋白的核心中有22个高风险nsSNP。预计这些高风险的致病性nsSNP会导致氨基酸的物理和化学性质发生变化,导致蛋白质的结构变化以及结构域与其他分子之间相互作用的变化,从而影响蛋白质的功能。该研究为缩小致病性CRYBA4 nsSNPs的数量和研究先天性白内障的发病机理提供了重要的参考价值。通过这种方法,我们可以轻松地从157个CRYBA4 nsSNP中找到44个高致病性nsSNP。



























 京公网安备 11010802027423号
京公网安备 11010802027423号