当前位置:
X-MOL 学术
›
J. Allergy Clin. Immunol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Lymphocyte DNA methylation mediates genetic risk at shared immune-mediated disease loci.
Journal of Allergy and Clinical Immunology ( IF 14.2 ) Pub Date : 2020-01-13 , DOI: 10.1016/j.jaci.2019.12.910 Alexander D Clark 1 , Nisha Nair 2 , Amy E Anderson 1 , Nishanthi Thalayasingam 1 , Najib Naamane 1 , Andrew J Skelton 3 , Julie Diboll 1 , Anne Barton 2 , Stephen Eyre 2 , John D Isaacs 4 , Arthur G Pratt 4 , Louise N Reynard 5
Journal of Allergy and Clinical Immunology ( IF 14.2 ) Pub Date : 2020-01-13 , DOI: 10.1016/j.jaci.2019.12.910 Alexander D Clark 1 , Nisha Nair 2 , Amy E Anderson 1 , Nishanthi Thalayasingam 1 , Najib Naamane 1 , Andrew J Skelton 3 , Julie Diboll 1 , Anne Barton 2 , Stephen Eyre 2 , John D Isaacs 4 , Arthur G Pratt 4 , Louise N Reynard 5
Affiliation
|
BACKGROUND
Defining regulatory mechanisms through which noncoding risk variants influence the cell-mediated pathogenesis of immune-mediated disease (IMD) has emerged as a priority in the post-genome-wide association study era.
OBJECTIVES
With a focus on rheumatoid arthritis, we sought new insight into genetic mechanisms of adaptive immune dysregulation to help prioritize molecular pathways for targeting in this and related immune pathologies.
METHODS
Whole-genome methylation and transcriptional data from isolated CD4+ T cells and B cells of more than 100 genotyped and phenotyped patients with inflammatory arthritis, all of whom were naive to immunomodulatory treatments, were obtained. Analysis integrated these comprehensive data with genome-wide association study findings across IMDs and other publicly available resources.
RESULTS
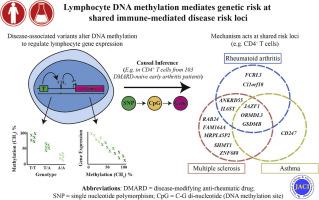
We provide strong evidence that disease-associated DNA variants regulate cis-CpG methylation in CD4+ T and/or B cells at 37% RA loci. Using paired, cell-specific transcriptomic data and causal inference testing, we identify examples where site-specific DNA methylation in turn mediates gene expression, including FCRL3 in both cell types and ORMDL3/GSDMB, IL6ST/ANKRD55, and JAZF1 in CD4+ T cells. A number of genes regulated in this way highlight mechanisms common to RA and other IMDs including multiple sclerosis and asthma, in turn distinguishing them from osteoarthritis, a primarily degenerative disease. Finally, we corroborate the observed effects experimentally.
CONCLUSIONS
Our observations highlight important mechanisms of genetic risk in RA and the wider context of immune dysregulation. They confirm the utility of DNA methylation profiling as a tool for causal gene prioritization and, potentially, therapeutic targeting in complex IMD.
中文翻译:

淋巴细胞 DNA 甲基化介导共享免疫介导疾病位点的遗传风险。
背景 在后全基因组关联研究时代,定义非编码风险变异影响细胞介导的免疫介导疾病 (IMD) 发病机制的调控机制已成为优先事项。目标 以类风湿性关节炎为重点,我们寻求对适应性免疫失调的遗传机制的新见解,以帮助优先考虑靶向这种和相关免疫病理学的分子途径。方法 获得来自 100 多名基因型和表型的炎症性关节炎患者的分离的 CD4+ T 细胞和 B 细胞的全基因组甲基化和转录数据,这些患者均未接受免疫调节治疗。分析将这些综合数据与跨 IMD 和其他公开可用资源的全基因组关联研究结果相结合。结果 我们提供了强有力的证据表明,疾病相关的 DNA 变体在 37% RA 基因座处调节 CD4+ T 和/或 B 细胞中的 cis-CpG 甲基化。使用配对的细胞特异性转录组数据和因果推理测试,我们确定了位点特异性 DNA 甲基化反过来介导基因表达的例子,包括两种细胞类型中的 FCRL3 和 CD4+ T 细胞中的 ORMDL3/GSDMB、IL6ST/ANKRD55 和 JAZF1。以这种方式调节的许多基因突出了 RA 和其他 IMD 的共同机制,包括多发性硬化症和哮喘,进而将它们与骨关节炎(一种主要的退行性疾病)区分开来。最后,我们通过实验证实了观察到的效果。结论 我们的观察强调了 RA 遗传风险的重要机制和免疫失调的更广泛背景。
更新日期:2020-01-13
中文翻译:

淋巴细胞 DNA 甲基化介导共享免疫介导疾病位点的遗传风险。
背景 在后全基因组关联研究时代,定义非编码风险变异影响细胞介导的免疫介导疾病 (IMD) 发病机制的调控机制已成为优先事项。目标 以类风湿性关节炎为重点,我们寻求对适应性免疫失调的遗传机制的新见解,以帮助优先考虑靶向这种和相关免疫病理学的分子途径。方法 获得来自 100 多名基因型和表型的炎症性关节炎患者的分离的 CD4+ T 细胞和 B 细胞的全基因组甲基化和转录数据,这些患者均未接受免疫调节治疗。分析将这些综合数据与跨 IMD 和其他公开可用资源的全基因组关联研究结果相结合。结果 我们提供了强有力的证据表明,疾病相关的 DNA 变体在 37% RA 基因座处调节 CD4+ T 和/或 B 细胞中的 cis-CpG 甲基化。使用配对的细胞特异性转录组数据和因果推理测试,我们确定了位点特异性 DNA 甲基化反过来介导基因表达的例子,包括两种细胞类型中的 FCRL3 和 CD4+ T 细胞中的 ORMDL3/GSDMB、IL6ST/ANKRD55 和 JAZF1。以这种方式调节的许多基因突出了 RA 和其他 IMD 的共同机制,包括多发性硬化症和哮喘,进而将它们与骨关节炎(一种主要的退行性疾病)区分开来。最后,我们通过实验证实了观察到的效果。结论 我们的观察强调了 RA 遗传风险的重要机制和免疫失调的更广泛背景。


























 京公网安备 11010802027423号
京公网安备 11010802027423号