当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Protein-Ligand Complex Solvation Thermodynamics: Development, Parameterization, and Testing of GIST-Based Solvent Functionals.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-01-10 , DOI: 10.1021/acs.jcim.9b01109 Tobias Hüfner-Wulsdorf 1 , Gerhard Klebe 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-01-10 , DOI: 10.1021/acs.jcim.9b01109 Tobias Hüfner-Wulsdorf 1 , Gerhard Klebe 1
Affiliation
|
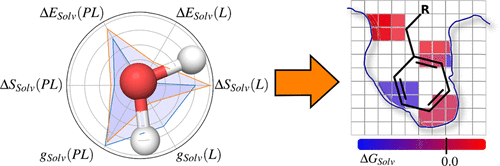
In drug design, the importance of molecular solvation and desolvation is increasingly appreciated and water molecules are recognized as active contributors to protein-ligand binding. However, despite a number of theoretical approaches, computational tools are still far from routinely integrating solvation features into rational structure-affinity relationships (SARs). In this contribution, we present a set of solvent functional-based models, which calculate the relative binding free energy contributions resulting from solvation for a diverse set of 53 thrombin protein-ligand complexes. These protein-ligand complexes were further matched into chemically similar pairs of ligand molecules. Our solvent functionals are based on molecular dynamics simulations in conjunction with grid inhomogeneous solvation theory (GIST) processing, and they are calibrated using accurate experimental data from isothermal titration calorimetry (ITC) measurements. We found that excellent agreement with experimental measurements can be achieved by considering either the desolvation of the protein-binding pocket or the ligand in solution prior to binding. The incorporation of contributions from the protein-ligand complexes generally results in good agreement with experimental measurements but require additional adjustment of spatial cutoff parameters. In addition, we investigated the transfer of the trained solvent functionals to another protein target, which revealed deviating performance results, indicating a target-specific treatment of solvation features within the model. Together with our tool GIST-based processing of solvent functionals (Gips), we provide a way to automatically generate solvent functional parameters from GIST data and allow for the design of compounds with favorable solvation properties given the chemical similarity and affinity range of the matching pairs in the training set. Finally, we reflect on the resemblance with the popular three-dimensional quantitative SAR (3D-QSAR) method, as our study allows for (retrospective) insights on the high predictive power of this well-established method.
中文翻译:

蛋白质-配体复杂溶剂化热力学:基于GIST的溶剂功能的开发,参数化和测试。
在药物设计中,人们越来越认识到分子溶剂化和去溶剂化的重要性,并且水分子被认为是蛋白质-配体结合的积极贡献者。但是,尽管有许多理论方法,但计算工具仍远远没有将溶剂化特征常规地整合到合理的结构亲和关系(SAR)中。在此贡献中,我们提出了一组基于溶剂功能的模型,该模型计算了由溶剂化形成的一组53种凝血酶蛋白-配体复合物的相对结合自由能的贡献。这些蛋白质-配体复合物进一步匹配成化学上相似的配体分子对。我们的溶剂功能基于分子动力学模拟以及网格非均质溶剂化理论(GIST)处理,并使用等温滴定量热(ITC)测量的准确实验数据对它们进行了校准。我们发现,通过考虑蛋白质结合口袋或配体在结合之前在溶液中的去溶剂化,可以实现与实验测量的极好的一致性。来自蛋白质-配体复合物的贡献的掺入通常导致与实验测量良好吻合,但是需要空间截止参数的额外调整。此外,我们调查了训练有素的溶剂功能转移到另一个蛋白质目标,这揭示了偏离的性能结果,表明模型中溶剂化特征的目标特异性处理。加上我们基于GIST的溶剂功能(Gips)处理工具,我们提供了一种从GIST数据自动生成溶剂功能参数的方法,并允许在给定训练集中匹配对的化学相似性和亲和力范围内的情况下,设计具有良好溶剂化性质的化合物。最后,我们反思与流行的三维定量SAR(3D-QSAR)方法的相似之处,因为我们的研究允许(回顾性)洞察这种成熟方法的高预测能力。
更新日期:2020-01-10
中文翻译:

蛋白质-配体复杂溶剂化热力学:基于GIST的溶剂功能的开发,参数化和测试。
在药物设计中,人们越来越认识到分子溶剂化和去溶剂化的重要性,并且水分子被认为是蛋白质-配体结合的积极贡献者。但是,尽管有许多理论方法,但计算工具仍远远没有将溶剂化特征常规地整合到合理的结构亲和关系(SAR)中。在此贡献中,我们提出了一组基于溶剂功能的模型,该模型计算了由溶剂化形成的一组53种凝血酶蛋白-配体复合物的相对结合自由能的贡献。这些蛋白质-配体复合物进一步匹配成化学上相似的配体分子对。我们的溶剂功能基于分子动力学模拟以及网格非均质溶剂化理论(GIST)处理,并使用等温滴定量热(ITC)测量的准确实验数据对它们进行了校准。我们发现,通过考虑蛋白质结合口袋或配体在结合之前在溶液中的去溶剂化,可以实现与实验测量的极好的一致性。来自蛋白质-配体复合物的贡献的掺入通常导致与实验测量良好吻合,但是需要空间截止参数的额外调整。此外,我们调查了训练有素的溶剂功能转移到另一个蛋白质目标,这揭示了偏离的性能结果,表明模型中溶剂化特征的目标特异性处理。加上我们基于GIST的溶剂功能(Gips)处理工具,我们提供了一种从GIST数据自动生成溶剂功能参数的方法,并允许在给定训练集中匹配对的化学相似性和亲和力范围内的情况下,设计具有良好溶剂化性质的化合物。最后,我们反思与流行的三维定量SAR(3D-QSAR)方法的相似之处,因为我们的研究允许(回顾性)洞察这种成熟方法的高预测能力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号