当前位置:
X-MOL 学术
›
Cancer Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
OSdlbcl: An online consensus survival analysis web server based on gene expression profiles of diffuse large B-cell lymphoma.
Cancer Medicine ( IF 4 ) Pub Date : 2020-01-09 , DOI: 10.1002/cam4.2829 Huan Dong 1 , Qiang Wang 1 , Guosen Zhang 1 , Ning Li 1 , Mengsi Yang 1 , Yang An 1 , Longxiang Xie 1 , Huimin Li 1 , Lu Zhang 1 , Wan Zhu 2 , Shuchun Zhao 3 , Haiyu Zhang 3 , Xiangqian Guo 1
Cancer Medicine ( IF 4 ) Pub Date : 2020-01-09 , DOI: 10.1002/cam4.2829 Huan Dong 1 , Qiang Wang 1 , Guosen Zhang 1 , Ning Li 1 , Mengsi Yang 1 , Yang An 1 , Longxiang Xie 1 , Huimin Li 1 , Lu Zhang 1 , Wan Zhu 2 , Shuchun Zhao 3 , Haiyu Zhang 3 , Xiangqian Guo 1
Affiliation
|
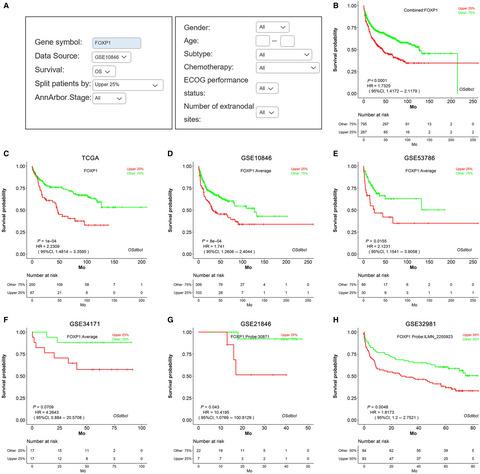
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of non-Hodgkin lymphoma (NHL) and is a clinical, pathological, and molecular heterogeneous disease with highly variable clinical outcomes. Currently, valid prognostic biomarkers in DLBCL are still lacking. To optimize targeted therapy and improve the prognosis of DLBCL, the performance of proposed biomarkers needs to be evaluated in multiple cohorts, and new biomarkers need to be investigated in large datasets. Here, we developed a consensus Online Survival analysis web server for Diffuse Large B-Cell Lymphoma, abbreviated OSdlbcl, to assess the prognostic value of individual gene. To build OSdlbcl, we collected 1100 samples with gene expression profiles and clinical follow-up information from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. In addition, DNA mutation data were also collected from the TCGA database. Overall survival (OS), progression-free survival (PFS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) are important endpoints to reflect the survival rate in OSdlbcl. Moreover, clinical features were integrated into OSdlbcl to allow data stratifications according to the user's special needs. By inputting an official gene symbol and selecting desired criteria, the survival analysis results can be graphically presented by the Kaplan-Meier (KM) plot with hazard ratio (HR) and log-rank p value. As a proof-of-concept demonstration, the prognostic value of 23 previously reported survival associated biomarkers, such as transcription factors FOXP1 and BCL2, was evaluated in OSdlbcl and found to be significantly associated with survival as reported (HR = 1.73, P < .01; HR = 1.47, P = .03, respectively). In conclusion, OSdlbcl is a new web server that integrates public gene expression, gene mutation data, and clinical follow-up information to provide prognosis evaluations for biomarker development for DLBCL. The OSdlbcl web server is available at https://bioinfo.henu.edu.cn/DLBCL/DLBCLList.jsp.
中文翻译:

OSdlbcl:基于弥散性大B细胞淋巴瘤的基因表达谱的在线共识生存分析Web服务器。
弥漫性大B细胞淋巴瘤(DLBCL)是非霍奇金淋巴瘤(NHL)的最常见亚型,是一种临床,病理和分子异质性疾病,临床结局变化很大。目前,仍然缺乏DLBCL中有效的预后生物标志物。为了优化靶向治疗并改善DLBCL的预后,需要在多个队列中评估提议的生物标志物的性能,并且需要在大型数据集中研究新的生物标志物。在这里,我们为弥漫性大型B细胞淋巴瘤(缩写为OSdlbcl)开发了一种共识在线生存分析Web服务器,以评估单个基因的预后价值。为了构建OSdlbcl,我们从The Cancer Genome Atlas(TCGA)和Gene Expression Omnibus(GEO)数据库中收集了1100个具有基因表达谱和临床随访信息的样本。此外,还从TCGA数据库收集了DNA突变数据。总生存期(OS),无进展生存期(PFS),疾病特异性生存期(DSS),无疾病间隔期(DFI)和无进展间隔期(PFI)是反映OSdlbcl生存率的重要终点。此外,将临床功能集成到OSdlbcl中,可以根据用户的特殊需求对数据进行分层。通过输入正式的基因符号并选择所需的标准,生存分析结果可以通过Kaplan-Meier(KM)图以危险比(HR)和对数p值图形表示。作为概念验证的证据,先前报道的23种与生存相关的生物标记物(例如转录因子FOXP1和BCL2,在OSdlbcl中进行了评估,发现与生存显着相关(如HR = 1.73,P <.01; HR = 1.47,P = .03)。总之,OSdlbcl是一个新的Web服务器,它集成了公共基因表达,基因突变数据和临床随访信息,从而为DLBCL的生物标志物开发提供了预后评估。OSdlbcl Web服务器可从https://bioinfo.henu.edu.cn/DLBCL/DLBCLList.jsp获得。
更新日期:2020-01-11
中文翻译:

OSdlbcl:基于弥散性大B细胞淋巴瘤的基因表达谱的在线共识生存分析Web服务器。
弥漫性大B细胞淋巴瘤(DLBCL)是非霍奇金淋巴瘤(NHL)的最常见亚型,是一种临床,病理和分子异质性疾病,临床结局变化很大。目前,仍然缺乏DLBCL中有效的预后生物标志物。为了优化靶向治疗并改善DLBCL的预后,需要在多个队列中评估提议的生物标志物的性能,并且需要在大型数据集中研究新的生物标志物。在这里,我们为弥漫性大型B细胞淋巴瘤(缩写为OSdlbcl)开发了一种共识在线生存分析Web服务器,以评估单个基因的预后价值。为了构建OSdlbcl,我们从The Cancer Genome Atlas(TCGA)和Gene Expression Omnibus(GEO)数据库中收集了1100个具有基因表达谱和临床随访信息的样本。此外,还从TCGA数据库收集了DNA突变数据。总生存期(OS),无进展生存期(PFS),疾病特异性生存期(DSS),无疾病间隔期(DFI)和无进展间隔期(PFI)是反映OSdlbcl生存率的重要终点。此外,将临床功能集成到OSdlbcl中,可以根据用户的特殊需求对数据进行分层。通过输入正式的基因符号并选择所需的标准,生存分析结果可以通过Kaplan-Meier(KM)图以危险比(HR)和对数p值图形表示。作为概念验证的证据,先前报道的23种与生存相关的生物标记物(例如转录因子FOXP1和BCL2,在OSdlbcl中进行了评估,发现与生存显着相关(如HR = 1.73,P <.01; HR = 1.47,P = .03)。总之,OSdlbcl是一个新的Web服务器,它集成了公共基因表达,基因突变数据和临床随访信息,从而为DLBCL的生物标志物开发提供了预后评估。OSdlbcl Web服务器可从https://bioinfo.henu.edu.cn/DLBCL/DLBCLList.jsp获得。


























 京公网安备 11010802027423号
京公网安备 11010802027423号