当前位置:
X-MOL 学术
›
Commun. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations.
Communications Biology ( IF 5.9 ) Pub Date : 2020-01-10 , DOI: 10.1038/s42003-019-0748-0 C Keith Cassidy 1, 2 , Benjamin A Himes 3 , Dapeng Sun 3 , Jun Ma 3 , Gongpu Zhao 3 , John S Parkinson 4 , Phillip J Stansfeld 1, 5 , Zaida Luthey-Schulten 2, 6 , Peijun Zhang 3, 7, 8
Communications Biology ( IF 5.9 ) Pub Date : 2020-01-10 , DOI: 10.1038/s42003-019-0748-0 C Keith Cassidy 1, 2 , Benjamin A Himes 3 , Dapeng Sun 3 , Jun Ma 3 , Gongpu Zhao 3 , John S Parkinson 4 , Phillip J Stansfeld 1, 5 , Zaida Luthey-Schulten 2, 6 , Peijun Zhang 3, 7, 8
Affiliation
|
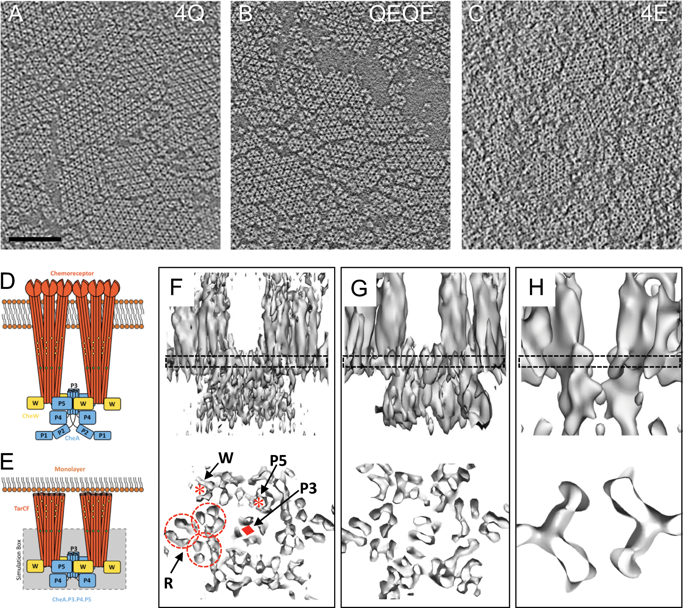
To enable the processing of chemical gradients, chemotactic bacteria possess large arrays of transmembrane chemoreceptors, the histidine kinase CheA, and the adaptor protein CheW, organized as coupled core-signaling units (CSU). Despite decades of study, important questions surrounding the molecular mechanisms of sensory signal transduction remain unresolved, owing especially to the lack of a high-resolution CSU structure. Here, we use cryo-electron tomography and sub-tomogram averaging to determine a structure of the Escherichia coli CSU at sub-nanometer resolution. Based on our experimental data, we use molecular simulations to construct an atomistic model of the CSU, enabling a detailed characterization of CheA conformational dynamics in its native structural context. We identify multiple, distinct conformations of the critical P4 domain as well as asymmetries in the localization of the P3 bundle, offering several novel insights into the CheA signaling mechanism.
中文翻译:

大肠杆菌趋化性核心信号复合物的结构和动力学通过低温电子层析成像和分子模拟。
为了能够处理化学梯度,趋化细菌拥有大量的跨膜化学感受器,组氨酸激酶CheA和衔接蛋白CheW,它们被组织为耦合核心信号单元(CSU)。尽管进行了数十年的研究,但由于缺乏高分辨率的CSU结构,围绕传感信号转导的分子机制的重要问题仍未解决。在这里,我们使用低温电子断层扫描和平均子断层图来确定亚纳米分辨率下的大肠杆菌CSU的结构。根据我们的实验数据,我们使用分子模拟来构建CSU的原子模型,从而在其天然结构环境下对CheA构象动力学进行详细表征。我们确定多个
更新日期:2020-01-10
中文翻译:

大肠杆菌趋化性核心信号复合物的结构和动力学通过低温电子层析成像和分子模拟。
为了能够处理化学梯度,趋化细菌拥有大量的跨膜化学感受器,组氨酸激酶CheA和衔接蛋白CheW,它们被组织为耦合核心信号单元(CSU)。尽管进行了数十年的研究,但由于缺乏高分辨率的CSU结构,围绕传感信号转导的分子机制的重要问题仍未解决。在这里,我们使用低温电子断层扫描和平均子断层图来确定亚纳米分辨率下的大肠杆菌CSU的结构。根据我们的实验数据,我们使用分子模拟来构建CSU的原子模型,从而在其天然结构环境下对CheA构象动力学进行详细表征。我们确定多个



























 京公网安备 11010802027423号
京公网安备 11010802027423号