Nature Methods ( IF 48.0 ) Pub Date : 2020-01-06 , DOI: 10.1038/s41592-019-0690-6 Aditya Pratapa 1 , Amogh P Jalihal 2 , Jeffrey N Law 2 , Aditya Bharadwaj 1 , T M Murali 1
|
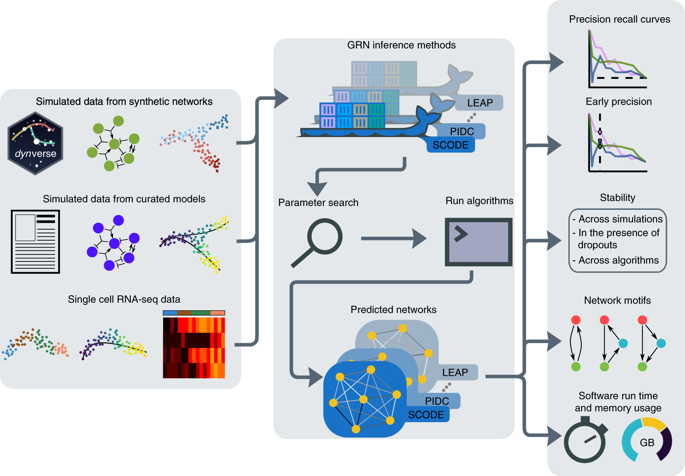
We present a systematic evaluation of state-of-the-art algorithms for inferring gene regulatory networks from single-cell transcriptional data. As the ground truth for assessing accuracy, we use synthetic networks with predictable trajectories, literature-curated Boolean models and diverse transcriptional regulatory networks. We develop a strategy to simulate single-cell transcriptional data from synthetic and Boolean networks that avoids pitfalls of previously used methods. Furthermore, we collect networks from multiple experimental single-cell RNA-seq datasets. We develop an evaluation framework called BEELINE. We find that the area under the precision-recall curve and early precision of the algorithms are moderate. The methods are better in recovering interactions in synthetic networks than Boolean models. The algorithms with the best early precision values for Boolean models also perform well on experimental datasets. Techniques that do not require pseudotime-ordered cells are generally more accurate. Based on these results, we present recommendations to end users. BEELINE will aid the development of gene regulatory network inference algorithms.
中文翻译:

从单细胞转录组数据推断基因调控网络的基准算法
我们对用于从单细胞转录数据推断基因调控网络的最先进算法进行了系统评估。作为评估准确性的基础事实,我们使用具有可预测轨迹的合成网络、文献整理的布尔模型和不同的转录调控网络。我们开发了一种策略来模拟来自合成和布尔网络的单细胞转录数据,以避免以前使用的方法的陷阱。此外,我们从多个实验性单细胞 RNA-seq 数据集中收集网络。我们开发了一个名为 BEELINE 的评估框架。我们发现算法的精确召回曲线下面积和早期精确度适中。这些方法在恢复合成网络中的交互方面比布尔模型更好。布尔模型具有最佳早期精度值的算法在实验数据集上也表现良好。不需要伪时间排序单元的技术通常更准确。基于这些结果,我们向最终用户提出建议。BEELINE 将帮助开发基因调控网络推理算法。



























 京公网安备 11010802027423号
京公网安备 11010802027423号