当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Enzymatic Synthesis of Designer DNA Using Cyclic Reversible Termination and a Universal Template.
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-01-14 , DOI: 10.1021/acssynbio.9b00315 Kendall Hoff 1 , Michelle Halpain 1 , Giancarlo Garbagnati 1 , Jeremy S Edwards 1, 2, 3, 4 , Wei Zhou 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-01-14 , DOI: 10.1021/acssynbio.9b00315 Kendall Hoff 1 , Michelle Halpain 1 , Giancarlo Garbagnati 1 , Jeremy S Edwards 1, 2, 3, 4 , Wei Zhou 1
Affiliation
|
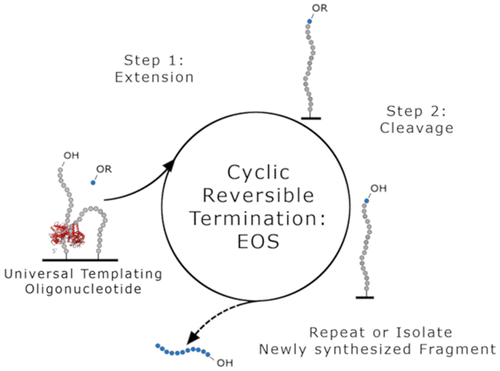
Phosphoramidite chemistry remains the industry standard for DNA synthesis despite significant limitations on the length and yield of the oligonucleotide, time restrictions, and hazardous waste production. Herein, we demonstrate the synthesis of single-stranded oligos on a solid surface by DNA polymerases and reverse transcriptases. We report the extension of surface-bound oligonucleotides enabled by transient hybridization of as few as two bases to a neighboring strand. When multiple hybridization structures are possible, each templating a different base, a DNA polymerase or reverse transcriptase can extend the oligonucleotide with any of the complementary bases. Therefore, the sequence of the newly synthesized fragment can be controlled by adding only the desired base as a substrate to the reaction solution. We used this enzymatic approach to synthesize a 20 base oligonucleotide by incorporating reversible terminator dNTPs through a two-step cyclic reversible termination process with a corrected stepwise efficiency over 98%. In our approach, a nascent DNA strand that serves as both primer and template is extended through polymerase-controlled sequential addition of 3'-reversibly blocked nucleotides followed by subsequent cleavage of the 3'-capping group. This process enables oligonucleotide synthesis in an environment not permitted by traditional phosphoramidite methods, eliminates the need for hazardous chemicals, has the potential to provide faster and higher yield results, and synthesizes DNA on a solid support with a free 3' end.
中文翻译:

使用循环可逆终止和通用模板酶促合成设计师DNA。
亚磷酰胺化学仍然是DNA合成的工业标准,尽管在寡核苷酸的长度和产量,时间限制和有害废物产生方面存在重大限制。在本文中,我们证明了通过DNA聚合酶和逆转录酶在固体表面上合成单链寡核苷酸。我们报道了通过少至两个碱基到相邻链的瞬时杂交而实现的表面结合寡核苷酸的延伸。当可能有多个杂交结构时,每个模板都具有不同的碱基,DNA聚合酶或逆转录酶可以使寡核苷酸与任何互补碱基一起延伸。因此,可以通过仅将期望的碱基作为底物添加到反应溶液中来控制新合成的片段的序列。我们使用这种酶促方法,通过两步循环可逆终止过程并以超过98%的修正逐步效率将可逆终止子dNTP掺入,从而合成了20个碱基的寡核苷酸。在我们的方法中,既可以用作引物又可以用作模板的新生DNA链,是通过聚合酶控制的顺序添加3'-可逆性封闭的核苷酸,然后随后切割3'-封端基团来扩展的。该方法能够在传统的亚磷酰胺方法所不允许的环境中进行寡核苷酸合成,消除了对有害化学物质的需求,具有提供更快和更高产量的潜力,并可以在具有游离3'端的固体支持物上合成DNA。
更新日期:2020-01-15
中文翻译:

使用循环可逆终止和通用模板酶促合成设计师DNA。
亚磷酰胺化学仍然是DNA合成的工业标准,尽管在寡核苷酸的长度和产量,时间限制和有害废物产生方面存在重大限制。在本文中,我们证明了通过DNA聚合酶和逆转录酶在固体表面上合成单链寡核苷酸。我们报道了通过少至两个碱基到相邻链的瞬时杂交而实现的表面结合寡核苷酸的延伸。当可能有多个杂交结构时,每个模板都具有不同的碱基,DNA聚合酶或逆转录酶可以使寡核苷酸与任何互补碱基一起延伸。因此,可以通过仅将期望的碱基作为底物添加到反应溶液中来控制新合成的片段的序列。我们使用这种酶促方法,通过两步循环可逆终止过程并以超过98%的修正逐步效率将可逆终止子dNTP掺入,从而合成了20个碱基的寡核苷酸。在我们的方法中,既可以用作引物又可以用作模板的新生DNA链,是通过聚合酶控制的顺序添加3'-可逆性封闭的核苷酸,然后随后切割3'-封端基团来扩展的。该方法能够在传统的亚磷酰胺方法所不允许的环境中进行寡核苷酸合成,消除了对有害化学物质的需求,具有提供更快和更高产量的潜力,并可以在具有游离3'端的固体支持物上合成DNA。



























 京公网安备 11010802027423号
京公网安备 11010802027423号