Abstract
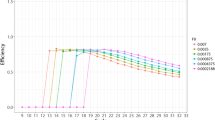
The experimental design that will be carried out to evaluate a nucleic acid quantification hypothesis determines the cost and feasibility of digital polymerase chain reaction (digital PCR) studies. Experiment design involves the calculation of the number of technical measurement replicates and the determination of the characteristics of those replicates, and this in accordance with the capabilities of the available digital PCR platform. Available digital PCR power analyses suffer from one or more of the following limitations: narrow scope, unrealistic assumptions, no sufficient detail for replication, lack of source code and user-friendly software. Here, we discuss the nature of six parameters that affect the statistical power, i.e., desired effect size, total number of partitions, fraction of positive partitions, number of replicate measurements, between-replicate variance, and significance level. We also show to what extent these parameters affect power, and argue that careful design of experiments is needed to achieve the desired power. A web tool, dPowerCalcR, that allows interactive calculation of statistical power and optimization of the experimental design is available.




Similar content being viewed by others
References
Baker M. Digital PCR hits its stride. Nat Methods 2012;9(6):541–4.
Hall Sedlak R, Jerome KR. The potential advantages of digital PCR for clinical virology diagnostics. Expert Rev Mol Diagn 2014;14(4):501–7.
Morisset D, Štebih D, Milavec M, Gruden K, žel J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One 2013;8(5):e62583.
Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital pcr. J Virol Methods 2012;186(1):68–72.
Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 2014;9(1):e85999.
Whale AS, Bushell CA, Grant PR, Cowen S, Gutierrez-Aguirre I, O’Sullivan DM, Zel J, Milavec M, Foy CA, Nastouli E. Detection of rare drug resistance mutations by digital PCR in a human influenza A virus model system and clinical samples. J Clin Microbiol 2016;54(2):392–400.
El Khattabi LA, Rouillac-Le Sciellour C, Le Tessier D, Luscan A, Coustier A, Porcher R, Bhouri R, Nectoux J, Sérazin V, Quibel T. Could digital PCR be an alternative as a non-invasive prenatal test for trisomy 21: a proof of concept study. PLoS One 2016;11(5):e0155009.
Colquhoun D. An investigation of the false discovery rate and the misinterpretation of p-values. Roy Soc Open Sci 2014;1(3):140216.
Whale AS, Huggett JF, Cowen S, Speirs V, Shaw J, Ellison S, Foy CA, Scott DJ. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res 2012;40(11):e82.
Evans MI, Wright DA, Pergament E, Cuckle HS, Nicolaides KH. Digital PCR for noninvasive detection of aneuploidy: power analysis equations for feasibility. Fetal Diagn Ther 2012;31(4):244–7.
Jones M, Williams J, Gärtner K, Phillips R, Hurst J, Frater J. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline,‘definetherain’. J Virol Methods 2014;202:46–53.
Lievens A, Jacchia S, Kagkli D, Savini C, Querci M. Measuring digital PCR quality: performance parameters and their optimization. PLoS One 2016;11(5):e0153317.
Vynck M, Vandesompele J, Nijs N, Menten B, De Ganck A, Thas O. Flexible analysis of digital PCR experiments using generalized linear mixed models. Biomol Detect Quantif 2016;9:1–13.
Trypsteen W, Vynck M, De Neve J, Bonczkowski P, Kiselinova M, Malatinkova E, Vervisch K, Thas O, Vandekerckhove L, De Spiegelaere W. ddpcRquant: threshold determination for single channel droplet digital PCR experiments. Anal Bioanal Chem 2015;407(19):5827–34.
Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Analytical chemistry 2011;84(2):1003–11.
Corbisier P, Pinheiro L, Mazoua S, Kortekaas AM, Chung PY, Gerganova T, Roebben G, Emons H, Emslie K. DNA Copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Analytical Bioanal Chem 2015;407(7):1831–40.
Vynck M, Thas O. Reducing bias in digital PCR quantification experiments: the importance of appropriately modeling volume variability. Anal Chem 2018;90(11):6540–7.
2018. R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing Vienna Austria. https://www.R-project.org/.
Jacobs BK, Goetghebeur E, Clement L. Impact of variance components on reliability of absolute quantification using digital PCR. BMC Bioinforma 2014;15(1):283.
Majumdar N, Wessel T, Marks J. Digital PCR modeling for maximal sensitivity dynamic range and measurement precision. PLoS One 2015;10(3):e0118833.
Huggett JF, Garson JA, Whale AS. Digital PCR and its potential application to microbiology. Molecular microbiology: diagnostic principles and practice. In: Persing DH, Tenover FC, Hayden RT, Ieven M, Miller MB, Nolte FS, Tang Y, and van Belkum A, editors. Washington, DC: ASM Press; 2016. p. 49–57.
Vynck M, Trypsteen W, Thas O, Vandekerckhove L, De Spiegelaere W. The future of digital polymerase chain reaction in virology. Mol Diagn Ther 2016;20(5):437–47.
Noble WS. How does multiple testing correction work? Nat Biotechnol 2000;27(12):1135–7.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 1995;57(1):289–300.
Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat 1979;6(2):65–70.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Biogazelle provided support in the form of salaries for JV, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. MV and OT have no conflict of interest to declare.
Additional information
Data availability
All data used is publicly available and referenced to in the main text. All code needed to reproduce our analyses is available at https://github.com/CenterForStatistics-UGent/dPowerCalcR.
Supplementary material
Electronic Supplementary Material (ESM) contains additional details on the derivation of the power equations, the sources of variation, the contribution of different sources of variation, additional figures of power curves for some specific designs, and the R/Shiny application.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vynck, M., Vandesompele, J. & Thas, O. On determining the power of digital PCR experiments. Anal Bioanal Chem 410, 5731–5739 (2018). https://doi.org/10.1007/s00216-018-1212-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1212-6