Abstract
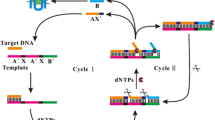
The authors describe a fluorometric method for the quantitation of nucleic acids by combining (a) cycled strand displacement amplification, (b) the unique features of the DNA probe SYBR Green, and (c) polydopamine nanotubes. SYBR Green undergoes strong fluorescence enhancement upon intercalation into double-stranded DNA (dsDNA). The polydopamine nanotubes selectively adsorb single-stranded DNA (ssDNA) and molecular beacons. In the absence of target DNA, the molecular beacon, primer and SYBR Green are adsorbed on the surface of polydopamine nanotubes. This results in quenching of the fluorescence of SYBR Green, typically measured at excitation/emission wavelengths of 488/518 nm. Upon addition of analyte (target DNA) and polymerase, the stem of the molecular beacon is opened so that it can bind to the primer. This triggers target strand displacement polymerization, during which dsDNA is synthesized. The hybridized target is then displaced due to the strand displacement activity of the polymerase. The displaced target hybridizes with another molecular beacon. This triggers the next round of polymerization. Consequently, a large amount of dsDNA is formed which is detected by addition of SYBR Green. Thus, sensitive and selective fluorometric detection is realized. The fluorescent sensing strategy shows very good analytical performances towards DNA detection, such as a wide linear range from 0.05 to 25 nM with a low limit of detection of 20 pM.

Schematic of a fluorometric strategy for highly sensitive and selective determination of nucleic acids by combining strand displacement amplification and the unique features of SYBR Green I (SG) and polydopamine nanotubes.




Similar content being viewed by others
References
Liu JW, Cao ZH, Lu Y (2009) Functional nucleic acid sensors. Chem Rev 109:1948–1998
Hang Y, Zhang YL, Xu XM, Jiang JH, Shen GL, Yu RQ (2009) Highly specific and sensitive electrochemical genotyping via gap ligation reaction and surface hybridization detection. J Am Chem Soc 131:2478–2480
Zheng J, Jiao AL, Yang RH, Li HM, Li JS, Shi ML, Ma C, Jiang Y, Deng L, Tan WH (2012) Fabricating a reversible and regenerable Raman-active substrate with a biomolecule-controlled DNA nanomachine. J Am Chem Soc 134:19957–19960
Zhao JJ, Chu ZD, Jing X, Zhao SL (2015) A fluorescence polarization assay for nucleic acid based on theamplification of hybridization chain reaction and nanoparticles. Sensors Actuators B Chem 209:116–121
Ma DL, He HZ, Leung KH, Zhong HJ, Chan DS, Leung CH (2013) Label-free luminescent oligonucleotide-based probes. Chem Soc Rev 42:3427–3440
Yin D, Tao YY, Tang L, Li W, Zhang Z, Li JL, Xie GM (2017) Cascade toehold-mediated strand displacement along with non-enzymatic target recycling amplification for the electrochemical determination of the HIV-1 related gene. Microchim Acta 184:3721–3728
Wang XU, Liu WW, Yin BB, Sang YW, Liu ZP, Dai Y, Duan XZ, Zhang G, Ding SJ, Tao ZH (2017) An isothermal strand displacement amplification strategy for nucleic acids using junction forming probes and colorimetric detection. Microchim Acta 184:1603–1610
Lee HJ, Li Y, Wark AW, Corn RW (2005) Enzymatically amplified surface plasmon resonance imaging detection of DNA by exonuclease III digestion of DNA microarrays. Anal Chem 77:5096–5100
Shi H, He XX, Yuan Y, Wang KM, Liu D (2010) Nanoparticle-based biocompatible and long-life marker for lysosome labeling and tracking. Anal Chem 82:2213–2220
FZ X, Shi H, He XX, Wang KM, He DG, Guo QP, Qing ZH, Yan LA, Ye XS, Li D, Tang JL (2014) Concatemeric dsDNA-templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in microRNA detection. Anal Chem 86:6976–6982
Ge J, Geng X, YH D, Chen JJ, Zhang L, Bai DM, Ji DY, YL H, Li ZH (2017) Highly sensitive fluorescence detection of mercury (II) ions based on WS2 nanosheets and T7 exonuclease assisted cyclic enzymatic amplification. Sensors Actuators B Chem 249:189–194
Wang Y, Li ZH, DH H, Lin CT, Li JH, Lin YH (2010) Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J Am Chem Soc 132:9274–9276
Gao C, Guo Z, Liu JH, Huang XJ (2012) The new age of carbon nanotubes: an updated review of functionalized carbon nanotubes in electrochemical sensors. Nano 4:1948–1963
Yan LA, Shi H, He XX, Wang KM, Tang JL, Chen M, Ye XS, FZ X, Lei YL (2014) A versatile activatable fluorescence probing platform for cancer cells in vitro and in vivo based on self-assembled aptamer/carbon nanotube ensembles. Anal Chem 86:9271–9277
Macak JM, Zlamal M, Krysa J, Schmuki P (2007) Self-organized TiO2 nanotube layers as highly efficient photocatalysts. Small 3:300–304
Ding W, Wada M, Kameta N, Minamikawa H, Shimizu T, Masuda M (2011) Functionalized organic nanotubes as tubular nonviral gene transfer vector. J Control Release 156:70–75
Liu YL, Ai KL, LH L (2014) Polydopamine and its derivative materials: synthesis and promising applications in energy, environmental, and biomedical fields. Chem Rev 114:5057–5115
Yu X, Fan HL, Wang L, Jin ZX (2014) Formation of polydopamine nanofibers with the aid of folic acid. Angew Chem Int Ed 53:12600–12604
Lynge ME, van der Westen R, Postma A, Städler B (2011) Polydopamine-a nature-inspired polymer coating for biomedical science. Nano 3:4916–4928
Lin LS, Cong ZX, Cao JB, Ke KM, Peng QL, Gao JH, Yang HH, Liu G, Chen XY (2014) Multifunctional Fe3O4@polydopamine core-shell nanocomposites for intracellular mRNA detection and imaging-guided photothermal therapy. ACS Nano 8:3876–3883
Park J, Brust TF, Lee HJ, Lee SC, Watts VJ, Yeo Y (2014) Polydopamine-based simple and versatile surface modification of polymeric nano drug carriers. ACS Nano 8:3347–3356
Fan DQ, Zhu XQ, Zhai QF, Wang EK, Dong SJ (2016) Polydopamine nanotubes as an effective fluorescent quencher for highly sensitive and selective detection of biomolecules assisted with exonuclease III amplification. Anal Chem 88:9158–9165
Zhuang J, Fu L, Xu M, Yang H, Chen G, Tang D (2013) Sensitive electrochemical monitoring of nucleic acids coupling DNA nanostructures with hybridization chain reaction. Anal Chim Acta 783:17–23
Xiong E, Zhang X, Liu Y, Zhou J, Yu P, Li X, Chen J (2015) Ultrasensitive electrochemical detection of nucleic acids based on the dual-signaling electrochemical ratiometric method and exonuclease III-assisted target recycling amplification strategy. Anal Chem 87:7291–7296
Sun AL, Zhang YF, Wang XN (2015) Sensitive voltammetric determination of DNA via a target-induced strand-displacement reaction using quantum dot-labeled probe DNA. Microchim Acta 182:1403–1610
Murakami T, Sumaoka J, Komiyama M (2009) Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res 37:e19
Li C, Li Y, Xu X, Wang X, Chen Y, Yang X, Liu F, Li N (2014) Fast and quantitative differentiation of single-base mismatched DNA by initial reaction rate of catalytic hairpin assembly. Biosens Bioelectron 60:57–63
Li YB, Li RM, Li Z, Zhang MJ, Ling LS (2017) Fluorometric determination of Simian virus 40 based on strand displacement amplification and triplex DNA using a molecular beacon probe with a guanine-rich fragment of the stem region. Microchim Acta 184:557–562
Wang YM, Liu JW, Duan LY, Liu SJ, Jiang JH (2017) Aptamer-based fluorometric determination of ATP by using target-cycling strand displacement amplification and copper nanoclusters. Microchim Acta 184:4183–4188
CH L, Li J, Liu JJ, Yang HH, Chen X, Chen GN (2010) Increasing the sensitivity and single-base mismatch selectivity of the molecular beacon using graphene oxide as the “nanoquencher”. Chem Eur J 16:4889–4894
Liu Y, Liao R, Wang H, Gong H, Chen CY, Chen XM, Cai CQ (2018) Accurate and sensitive fluorescence detection of DNA base on G-quadruplex hairpin DNA. Talanta 176:422–427
Wei W, Gao CY, Xiong YX, Zhang YJ, Liu SQ, YP P (2015) A fluorescence method for detection of DNA and DNA methylation based on graphene oxide and restriction endonuclease HpaII. Talanta 131:342–347
Zhao CQ, Wu L, Ren JS, XG Q (2011) A label-free fluorescent turn-on enzymatic amplification assay for DNA detection using ligand-responsive G-quadruplex formation. Chem Commun 47:5461–5463
Luo M, Chen X, Zhou GH, Xiang X, Chen L, Ji XH, He ZK (2012) Chemiluminescence biosensors for DNA detection using graphene oxide and a horseradish peroxidase-mimicking DNAzyme. Chem Commun 48:1126–1128
Bonnet G, Tyagi S, Libchaber A, Kramer FR (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc Natl Acad Sci U S A 96:6171–6176
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21505122, 21205108), the Startup Research Fund of Zhengzhou University (1511316004), and the Outstanding Young Talent Research Fund of Zhengzhou University (1521316003, 1421316038).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOC 8730 kb)
Rights and permissions
About this article
Cite this article
Ge, J., Bai, DM., -Geng, X. et al. Fluorometric determination of nucleic acids based on the use of polydopamine nanotubes and target-induced strand displacement amplification. Microchim Acta 185, 105 (2018). https://doi.org/10.1007/s00604-017-2632-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-017-2632-2