Abstract
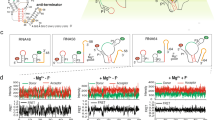
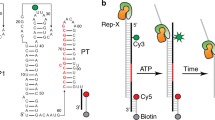
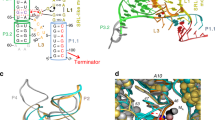
Riboswitches control gene expression through ligand-dependent structural rearrangements of the sensing aptamer domain. However, we found that the Bacillus cereus fluoride riboswitch aptamer adopts identical tertiary structures in solution with and without ligand. Using chemical-exchange saturation transfer (CEST) NMR spectroscopy, we revealed that the structured ligand-free aptamer transiently accesses a low-populated (∼1%) and short-lived (∼3 ms) excited conformational state that unravels a conserved 'linchpin' base pair to signal transcription termination. Upon fluoride binding, this highly localized, fleeting process is allosterically suppressed, which activates transcription. We demonstrated that this mechanism confers effective fluoride-dependent gene activation over a wide range of transcription rates, which is essential for robust toxicity responses across diverse cellular conditions. These results unveil a novel switching mechanism that employs ligand-dependent suppression of an aptamer excited state to coordinate regulatory conformational transitions rather than adopting distinct aptamer ground-state tertiary architectures, exemplifying a new mode of ligand-dependent RNA regulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Mironov, A.S. et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756 (2002).
Nahvi, A. et al. Genetic control by a metabolite binding mRNA. Chem. Biol. 9, 1043 (2002).
Winkler, W., Nahvi, A. & Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Serganov, A. & Nudler, E. A decade of riboswitches. Cell 152, 17–24 (2013).
Batey, R.T. Structure and mechanism of purine-binding riboswitches. Q. Rev. Biophys. 45, 345–381 (2012).
Noeske, J. et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. USA 102, 1372–1377 (2005).
Lee, M.K., Gal, M., Frydman, L. & Varani, G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. USA 107, 9192–9197 (2010).
Heppell, B. et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat. Chem. Biol. 7, 384–392 (2011).
Haller, A., Rieder, U., Aigner, M., Blanchard, S.C. & Micura, R. Conformational capture of the SAM-II riboswitch. Nat. Chem. Biol. 7, 393–400 (2011).
Wilson, R.C. et al. Tuning riboswitch regulation through conformational selection. J. Mol. Biol. 405, 926–938 (2011).
Frieda, K.L. & Block, S.M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400 (2012).
Chen, B., Zuo, X., Wang, Y.X. & Dayie, T.K. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res. 40, 3117–3130 (2012).
Suddala, K.C. et al. Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res. 41, 10462–10475 (2013).
Reining, A. et al. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature 499, 355–359 (2013).
Zhang, J., Jones, C.P. & Ferré-D'Amaré, A.R. Global analysis of riboswitches by small-angle X-ray scattering and calorimetry. Biochim. Biophys. Acta 1839, 1020–1029 (2014).
Ren, A. et al. Structural and dynamic basis for low-affinity, high-selectivity binding of L-glutamine by the glutamine riboswitch. Cell Rep. 13, 1800–1813 (2015).
Stoddard, C.D. et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18, 787–797 (2010).
Serganov, A., Huang, L. & Patel, D.J. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455, 1263–1267 (2008).
Huang, L., Serganov, A. & Patel, D.J. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol. Cell 40, 774–786 (2010).
Jenkins, J.L., Krucinska, J., McCarty, R.M., Bandarian, V. & Wedekind, J.E. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. J. Biol. Chem. 286, 24626–24637 (2011).
Vicens, Q., Mondragón, E. & Batey, R.T. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res. 39, 8586–8598 (2011).
Stagno, J.R. et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature 541, 242–246 (2017).
Baker, J.L. et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335, 233–235 (2012).
Li, S. et al. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc. Natl. Acad. Sci. USA 110, 19018–19023 (2013).
Ren, A., Rajashankar, K.R. & Patel, D.J. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486, 85–89 (2012).
Bothe, J.R. et al. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nat. Methods 8, 919–931 (2011).
Wickiser, J.K., Winkler, W.C., Breaker, R.R. & Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Sekhar, A. & Kay, L.E. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. USA 110, 12867–12874 (2013).
Palmer, A.G. III. Chemical exchange in biomacromolecules: past, present, and future. J. Magn. Reson. 241, 3–17 (2014).
Dethoff, E.A., Petzold, K., Chugh, J., Casiano-Negroni, A. & Al-Hashimi, H.M. Visualizing transient low-populated structures of RNA. Nature 491, 724–728 (2012).
Fawzi, N.L., Ying, J., Ghirlando, R., Torchia, D.A. & Clore, G.M. Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature 480, 268–272 (2011).
Vallurupalli, P., Bouvignies, G. & Kay, L.E. Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 (2012).
Zhao, B., Hansen, A.L. & Zhang, Q. Characterizing slow chemical exchange in nucleic acids by carbon CEST and low spin-lock field R(1ρ) NMR spectroscopy. J. Am. Chem. Soc. 136, 20–23 (2014).
Monforte, J.A., Kahn, J.D. & Hearst, J.E. RNA folding during transcription by Escherichia coli RNA polymerase analyzed by RNA self-cleavage. Biochemistry 29, 7882–7890 (1990).
Komissarova, N. & Kashlev, M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. USA 95, 14699–14704 (1998).
Gusarov, I. & Nudler, E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell 107, 437–449 (2001).
Yarnell, W.S. & Roberts, J.W. Mechanism of intrinsic transcription termination and antitermination. Science 284, 611–615 (1999).
Gusarov, I. & Nudler, E. The mechanism of intrinsic transcription termination. Mol. Cell 3, 495–504 (1999).
Abbondanzieri, E.A., Greenleaf, W.J., Shaevitz, J.W., Landick, R. & Block, S.M. Direct observation of base-pair stepping by RNA polymerase. Nature 438, 460–465 (2005).
Watters, K.E., Strobel, E.J., Yu, A.M., Lis, J.T. & Lucks, J.B. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat. Struct. Mol. Biol. 23, 1124–1131 (2016).
Wang, J.X., Lee, E.R., Morales, D.R., Lim, J. & Breaker, R.R. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell 29, 691–702 (2008).
Nelson, J.W., Atilho, R.M., Sherlock, M.E., Stockbridge, R.B. & Breaker, R.R. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol. Cell 65, 220–230 (2017).
Tzeng, S.R. & Kalodimos, C.G. Allosteric inhibition through suppression of transient conformational states. Nat. Chem. Biol. 9, 462–465 (2013).
Dethoff, E.A., Chugh, J., Mustoe, A.M. & Al-Hashimi, H.M. Functional complexity and regulation through RNA dynamics. Nature 482, 322–330 (2012).
Hoogstraten, C.G., Wank, J.R. & Pardi, A. Active site dynamics in the lead-dependent ribozyme. Biochemistry 39, 9951–9958 (2000).
Blad, H., Reiter, N.J., Abildgaard, F., Markley, J.L. & Butcher, S.E. Dynamics and metal ion binding in the U6 RNA intramolecular stem-loop as analyzed by NMR. J. Mol. Biol. 353, 540–555 (2005).
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Zhang, Q., Kim, N.K., Peterson, R.D., Wang, Z. & Feigon, J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc. Natl. Acad. Sci. USA 107, 18761–18768 (2010).
Duchardt-Ferner, E., Ferner, J. & Wöhnert, J. Rapid identification of noncanonical RNA structure elements by direct detection of OH⋯O=P, NH⋯O=P, and NH2⋯O=P hydrogen bonds in solution NMR spectroscopy. Angew. Chem. Int. Edn. Engl. 50, 7927–7930 (2011).
Bermejo, G.A., Clore, G.M. & Schwieters, C.D. Improving NMR Structures of RNA. Structure 24, 806–815 (2016).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 (1996).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Johnson, B.A. & Blevins, R.A. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994).
McConnell, H.M. Reaction rates by nuclear magnetic resonance. J. Chem. Phys. 28, 430–431 (1958).
Vallurupalli, P. & Kay, L.E. Probing slow chemical exchange at carbonyl sites in proteins by chemical exchange saturation transfer NMR spectroscopy. Angew. Chem. Int. Edn. Engl. 52, 4156–4159 (2013).
Landick, R., Wang, D. & Chan, C.L. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 274, 334–353 (1996).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Mejia, Y.X., Nudler, E. & Bustamante, C. Trigger loop folding determines transcription rate of Escherichia coli's RNA polymerase. Proc. Natl. Acad. Sci. USA 112, 743–748 (2015).
Xu, X., Yu, T. & Chen, S.J. Understanding the kinetic mechanism of RNA single base pair formation. Proc. Natl. Acad. Sci. USA 113, 116–121 (2016).
Acknowledgements
We thank G. Young for maintenance of NMR instruments and members of the Zhang lab for critical comments. This work was supported by start-up fund from the University of North Carolina at Chapel Hill and an NIH grant (R01 GM114432) to Q.Z.
Author information
Authors and Affiliations
Contributions
B.Z. and Q.Z. conceived the project and experimental design. B.Z. and Q.Z. prepared the samples, carried out NMR and biochemical experiments, analyzed the data, and wrote the paper. S.L.G., B.W., and Q.Z. analyzed NMR RDC data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–12 and Supplementary Table 1 (PDF 7656 kb)
Rights and permissions
About this article
Cite this article
Zhao, B., Guffy, S., Williams, B. et al. An excited state underlies gene regulation of a transcriptional riboswitch. Nat Chem Biol 13, 968–974 (2017). https://doi.org/10.1038/nchembio.2427
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2427
This article is cited by
-
An RNA excited conformational state at atomic resolution
Nature Communications (2023)
-
Observation of coordinated RNA folding events by systematic cotranscriptional RNA structure probing
Nature Communications (2023)
-
Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution
Nature Communications (2023)
-
Studying micro to millisecond protein dynamics using simple amide 15N CEST experiments supplemented with major-state R2 and visible peak-position constraints
Journal of Biomolecular NMR (2023)
-
Structure determination of high-energy states in a dynamic protein ensemble
Nature (2022)