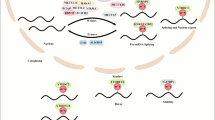
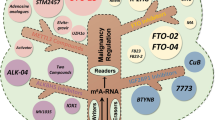
Abstract
Malignant liver cancer is characterized by rapid tumour progression and a high mortality rate, whereas the molecular mechanisms underlying liver cancer initiation and progression are still poorly understood. The dynamic and reversible RNA modifications have crucial functions in gene expression regulation by modulating RNA processing and mRNA translation. Emerging evidence has revealed that alterations in RNA modifications facilitate the selective translation of oncogenic transcripts and promote the diverse tumorigenic processes of liver cancer. In this Review, we first highlight the current progress on the functions and mechanisms underlying RNA modifications in the regulation of mRNA translation and then summarize the exciting discoveries on aberrant RNA modification-mediated mRNA translation in the regulation of tumour initiation, metastasis, metabolism, tumour microenvironment, and drug and radiotherapy resistance in liver cancer. Finally, we discuss the diagnostic and therapeutic potentials of targeting RNA modifications and mRNA translation for the clinical management of liver cancer.
Key points
-
Epigenetic changes, including DNA methylation, histone modifications and RNA modifications, have critical roles in liver cancer initiation and progression.
-
The dynamic and reversible RNA modifications have crucial functions in the regulation of RNA processing and mRNA translation.
-
Aberrant modifications on mRNAs, tRNAs and ribosomal RNAs regulate the translation of oncogenic transcripts and promote diverse tumorigenic aspects of liver cancer.
-
Targeting aberrant RNA modification-mediated mRNA translation could be promising for the development of effective diagnostic and therapeutic strategies for liver cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7, 6 (2021).
Rumgay, H. et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 77, 1598–1606 (2022).
Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462 (2019).
Hama, N. et al. Epigenetic landscape influences the liver cancer genome architecture. Nat. Commun. 9, 1643 (2018).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Zhao, Z. et al. Epitranscriptomics in liver disease: basic concepts and therapeutic potential. J. Hepatol. 73, 664–679 (2020).
Nusinow, D. P. et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell 180, 387–402.e16 (2020).
Bhat, M. et al. Targeting the translation machinery in cancer. Nat. Rev. Drug. Discov. 14, 261–278 (2015).
Frye, M., Harada, B. T., Behm, M. & He, C. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Wang, L. & Lin, S. Emerging functions of tRNA modifications in mRNA translation and diseases. J. Genet. Genomics 50, 223–232 (2022).
Orellana, E. A., Siegal, E. & Gregory, R. I. tRNA dysregulation and disease. Nat. Rev. Genet. 23, 651–664 (2022).
Sloan, K. E. et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152 (2017).
Yang, Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641 (2017).
Qian, B., Wang, P., Zhang, D. & Wu, L. m6A modification promotes miR-133a repression during cardiac development and hypertrophy via IGF2BP2. Cell Death Discov. 7, 157 (2021).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971–3975 (1974).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Yang, Y., Hsu, P. J., Chen, Y. S. & Yang, Y. G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 28, 616–624 (2018).
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Choe, J. et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018).
Meyer, KateD. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Coots, R. A. et al. m6A facilitates eIF4F-independent mRNA translation. Mol. Cell 68, 504–514.e7 (2017).
Choi, J. et al. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 23, 110–115 (2016).
Mao, Y. et al. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10, 5332 (2019).
Dominissini, D. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Li, X. et al. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e9 (2017).
Li, Q. et al. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J. Cell. Biochem. 118, 2587–2598 (2017).
Hoernes, T. P. et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44, 852–862 (2016).
Tang, H. et al. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging 7, 1143–1158 (2015).
Delatte, B. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Arango, D. et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24 (2018).
Arango, D. et al. Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol. Cell 82, 2797–2814.e11 (2022).
Malbec, L. et al. Dynamic methylome of internal mRNA N7-methylguanosine and its regulatory role in translation. Cell Res. 29, 927–941 (2019).
Zhang, L. S. et al. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 74, 1304–1316.e8 (2019).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Svitkin, Y. V. et al. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036 (2017).
Svitkin, Y. V., Gingras, A. C. & Sonenberg, N. Membrane-dependent relief of translation elongation arrest on pseudouridine- and N1-methyl-pseudouridine-modified mRNAs. Nucleic Acids Res. 50, 7202–7215 (2022).
Karijolich, J. & Yu, Y. T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398 (2011).
Hanson, G. & Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 (2018).
Schaffrath, R. & Leidel, S. A. Wobble uridine modifications – a reason to live, a reason to die?! RNA Biol. 14, 1209–1222 (2017).
Nguyen, H. A., Hoffer, E. D. & Dunham, C. M. Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing in tRNACGGPro for decoding. J. Biol. Chem. 294, 5281–5291 (2019).
Zhou, J. B., Wang, E. D. & Zhou, X. L. Modifications of the human tRNA anticodon loop and their associations with genetic diseases. Cell. Mol. Life Sci. 78, 7087–7105 (2021).
Lin, S. et al. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255.e5 (2018).
Liu, F. et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 167, 816–828.e6 (2016).
Liu, Y. et al. tRNA-m1A modification promotes T cell expansion via efficient MYC protein synthesis. Nat. Immunol. 23, 1433–1444 (2022).
Polikanov, Y. S., Melnikov, S. V., Söll, D. & Steitz, T. A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015).
Sharma, S. & Lafontaine, D. L. J. ‘View from a bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 40, 560–575 (2015).
Erales, J. et al. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl Acad. Sci. USA 114, 12934–12939 (2017).
Jack, K. et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44, 660–666 (2011).
Ma, H. et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94 (2019).
Peng, H. et al. N6-methyladenosine (m6A) in 18S rRNA promotes fatty acid metabolism and oncogenic transformation. Nat. Metab. 4, 1041–1054 (2022).
Haag, S., Kretschmer, J. & Bohnsack, M. T. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 21, 180–187 (2015).
Cámara, Y. et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 13, 527–539 (2011).
Barbieri, I. & Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322 (2020).
Chen, M. & Wong, C. M. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol. Cancer 19, 44 (2020).
Qi, L., Chan, T. H., Tenen, D. G. & Chen, L. RNA editome imbalance in hepatocellular carcinoma. Cancer Res. 74, 1301–1306 (2014).
Chen, M. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270 (2018).
Ma, J. Z. et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology 65, 529–543 (2017).
Zhang, Y. et al. The genomic landscape of cholangiocarcinoma reveals the disruption of post-transcriptional modifiers. Nat. Commun. 13, 3061 (2022).
Su, R. et al. METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 24, 205–216 (2022).
Dai, Y. Z. et al. METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m6A-dependent manner. Cell Mol. Biol. Lett. 27, 41 (2022).
Lan, T. et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 18, 186 (2019).
Cai, X. et al. RBM15 promotes hepatocellular carcinoma progression by regulating N6-methyladenosine modification of YES1 mRNA in an IGF2BP1-dependent manner. Cell Death Discov. 7, 315 (2021).
Chen, Y. et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 18, 127 (2019).
Ortiz-Barahona, V. et al. Epigenetic inactivation of the 5-methylcytosine RNA methyltransferase NSUN7 is associated with clinical outcome and therapeutic vulnerability in liver cancer. Mol. Cancer 22, 83 (2023).
Chen, Y. et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m6A-guided epigenetic inhibition of LYPD1. Mol. Cancer 19, 123 (2020).
Li, J. et al. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am. J. Transl. Res. 11, 6084–6092 (2019).
Liu, X. et al. SIRT1 regulates N6-methyladenosine RNA modification in hepatocarcinogenesis by inducing RANBP2-dependent FTO SUMOylation. Hepatology 72, 2029–2050 (2020).
Rong, Z. X. et al. Downregulation of fat mass and obesity associated (FTO) promotes the progression of intrahepatic cholangiocarcinoma. Front. Oncol. 9, 369 (2019).
Qin, S. et al. The comprehensive expression and functional analysis of m6A modification “readers” in hepatocellular carcinoma. Aging 14, 6269–6298 (2022).
Li, Q. et al. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal. Transduct. Target. Ther. 6, 76 (2021).
Zhong, L. et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 442, 252–261 (2019).
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Chen, Z. et al. METTL1 promotes hepatocarcinogenesis via m7G tRNA modification-dependent translation control. Clin. Transl. Med. 11, e661 (2021).
Dai, Z. et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell 81, 3339–3355.e8 (2021).
Tian, Q. H. et al. METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J. Mol. Med. 97, 1535–1545 (2019).
Wang, Y. et al. N1-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 12, 6314 (2021).
Ye, Y. et al. TRMT6 promotes hepatocellular carcinoma progression through the PI3K/AKT signaling pathway. Eur. J. Med. Res. 28, 48 (2023).
Ignatova, V. V. et al. METTL6 is a tRNA m3C methyltransferase that regulates pluripotency and tumor cell growth. Sci. Adv. 6, eaaz4551 (2020).
Liu, Z. et al. SNORA23 inhibits HCC tumorigenesis by impairing the 2′-O-ribose methylation level of 28S rRNA. Cancer Biol. Med. 19, 104–119 (2021).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014).
Lin, X. et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 10, 2065 (2019).
Xu, H. et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics 10, 5671–5686 (2020).
Li, J. et al. RNA m6A methylation regulates dissemination of cancer cells by modulating expression and membrane localization of β-catenin. Mol. Ther. : J. Am. Soc. Gene Ther. 30, 1578–1596 (2022).
Zhu, S. et al. Targeting N7-methylguanosine tRNA modification blocks hepatocellular carcinoma metastasis after insufficient radiofrequency ablation. Mol. Ther. 31, 1596–1614 (2023).
Fan, Z. et al. METTL3/IGF2BP1/CD47 contributes to the sublethal heat treatment induced mesenchymal transition in HCC. Biochem. Biophys. Res. Commun. 546, 169–177 (2021).
Luo, X., Cao, M., Gao, F. & He, X. YTHDF1 promotes hepatocellular carcinoma progression via activating PI3K/AKT/mTOR signaling pathway and inducing epithelial-mesenchymal transition. Exp. Hematol. Oncol. 10, 35 (2021).
Zhang, C. et al. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 39, 4507–4518 (2020).
Chen, Y. et al. Activation of YAP1 by N6-methyladenosine-modified circCPSF6 drives malignancy in hepatocellular carcinoma. Cancer Res. 82, 599–614 (2022).
Liu, L. et al. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellula\zar carcinoma. Mol. Cancer 21, 149 (2022).
Zhang, H. et al. ALKBH5-mediated m6A modification of lincRNA LINC02551 enhances the stability of DDX24 to promote hepatocellular carcinoma growth and metastasis. Cell Death Dis. 13, 926 (2022).
De Matteis, S. et al. Aberrant metabolism in hepatocellular carcinoma provides diagnostic and therapeutic opportunities. Oxid. Med. Cell. Longev. 2018, 7512159 (2018).
Shen, X. et al. The m6A methylation landscape stratifies hepatocellular carcinoma into 3 subtypes with distinct metabolic characteristics. Cancer Biol. Med. 17, 937–952 (2020).
Li, Z. et al. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 11, 2578 (2020).
Lin, Y., Wei, X., Jian, Z. & Zhang, X. METTL3 expression is associated with glycolysis metabolism and sensitivity to glycolytic stress in hepatocellular carcinoma. Cancer Med. 9, 2859–2867 (2020).
Gao, J. et al. Methyltransferase like 3 inhibition limits intrahepatic cholangiocarcinoma metabolic reprogramming and potentiates the efficacy of chemotherapy. Oncogene 42, 2507–2520 (2023).
Du, L. et al. USP48 is upregulated by Mettl14 to attenuate hepatocellular carcinoma via regulating SIRT6 stabilization. Cancer Res. 81, 3822–3834 (2021).
Zhao, L. et al. UBR7 inhibits HCC tumorigenesis by targeting Keap1/Nrf2/Bach1/HK2 and glycolysis. J. Exp. Clin. Cancer Res. 41, 330 (2022).
Zhou, R. et al. A functional loop between YTH domain family protein YTHDF3 mediated m6A modification and phosphofructokinase PFKL in glycolysis of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 41, 334 (2022).
Yang, Y. et al. Dysregulated m6A modification promotes lipogenesis and development of non-alcoholic fatty liver disease and hepatocellular carcinoma. Mol. Ther. 30, 2342–2353 (2022).
Zuo, X. et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 13, 5 (2020).
Ben-Haim, M. S. et al. Dynamic regulation of N6,2′-O-dimethyladenosine (m6Am) in obesity. Nat. Commun. 12, 7185 (2021).
Donne, R. & Lujambio, A. The liver cancer immune microenvironment: therapeutic implications for hepatocellular carcinoma. Hepatology 77, 1773–1796 (2022).
Zhou, D. et al. m6A regulator-mediated methylation modification highlights immune infiltration patterns for predicting risk in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 77, 3661–3680 (2022).
Qiu, X. et al. M6A demethylase ALKBH5 regulates PD-L1 expression and tumor immunoenvironment in intrahepatic cholangiocarcinoma. Cancer Res. 81, 4778–4793 (2021).
You, Y. et al. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int. J. Biol. Sci. 18, 5001–5018 (2022).
Hou, J. et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 18, 163 (2019).
Liu, Y. et al. Allosteric regulation of IGF2BP1 as a novel strategy for the activation of tumor immune microenvironment. ACS Cent. Sci. 8, 1102–1115 (2022).
Liu, H. et al. Targeting tumour-intrinsic N7-methylguanosine tRNA modification inhibits MDSC recruitment and improves anti-PD-1 efficacy. Gut 72, 1555–1567 (2022).
Zeng, X. et al. Eliminating METTL1-mediated accumulation of PMN-MDSCs prevents hepatocellular carcinoma recurrence after radiofrequency ablation. Hepatology 77, 1122–1138 (2022).
Kim, G. W. et al. HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology 73, 533–547 (2021).
Jin, Z. et al. Integrative multiomics evaluation reveals the importance of pseudouridine synthases in hepatocellular carcinoma. Front. Genet. 13, 944681 (2022).
Li, D. et al. The m6A/m5C/m1A regulated gene signature predicts the prognosis and correlates with the immune status of hepatocellular carcinoma. Front. Immunol. 13, 918140 (2022).
Chu, K. F. & Dupuy, D. E. Thermal ablation of tumours: biological mechanisms and advances in therapy. Nat. Rev. Cancer 14, 199–208 (2014).
Su, T. et al. Insufficient radiofrequency ablation promotes hepatocellular carcinoma metastasis through N6-methyladenosine mRNA methylation-dependent mechanism. Hepatology 74, 1339–1356 (2021).
Alexandrov, A., Grayhack, E. J. & Phizicky, E. M. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 11, 821–830 (2005).
Alexandrov, A. et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96 (2006).
Ohri, N. et al. Radiotherapy for hepatocellular carcinoma: new indications and directions for future study. J. Natl Cancer Inst. 108, djw133 (2016).
Xiang, M., Liu, W., Tian, W., You, A. & Deng, D. RNA N-6-methyladenosine enzymes and resistance of cancer cells to chemotherapy and radiotherapy. Epigenomics 12, 801–809 (2020).
Chen, Y. et al. ALKBH5-mediated m6A demethylation of TIRAP mRNA promotes radiation-induced liver fibrosis and decreases radiosensitivity of hepatocellular carcinoma. Clin. Transl. Med. 13, e1198 (2023).
Liao, J. et al. Methyltransferase 1 is required for nonhomologous end-joining repair and renders hepatocellular carcinoma resistant to radiotherapy. Hepatology 12, 1896–1910 (2022).
Yang, C. et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 20, 203–222 (2023).
Tang, W. et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal. Transduct. Target. Ther. 5, 87 (2020).
Lin, Z. et al. RNA m6A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 39, e103181 (2020).
Zhou, T. et al. m6A RNA methylation-mediated HNF3γ reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal. Transduct. Target. Ther. 5, 296 (2020).
Xu, J. et al. N6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol. Cancer 19, 163 (2020).
Wang, L. et al. METTL3-m6A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. 559, 216122 (2023).
Huang, M. et al. METTL1-mediated m7G tRNA modification promotes lenvatinib resistance in hepatocellular carcinoma. Cancer Res. 83, 89–102 (2023).
Jin, H. et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature 595, 730–734 (2021).
Ke, W., Zhang, L., Zhao, X. & Lu, Z. p53 m6A modulation sensitizes hepatocellular carcinoma to apatinib through apoptosis. Apoptosis 27, 426–440 (2022).
Castaldo, G. et al. Total discrimination of peritoneal malignant ascites from cirrhosis- and hepatocarcinoma-associated ascites by assays of ascitic cholesterol and lactate dehydrogenase. Clin. Chem. 40, 478–483 (1994).
Amuro, Y. et al. Serum pseudouridine as a biochemical marker in patients with hepatocellular carcinoma. Clin. Chim. Acta 178, 151–158 (1988).
Tamura, S. et al. Urinary pseudouridine as a biochemical marker in the diagnosis and monitoring of primary hepatocellular carcinoma. Am. J. Gastroenterol. 83, 841–845 (1988).
Zhang, B. et al. m6A target microRNAs in serum for cancer detection. Mol. Cancer 20, 170 (2021).
Liu, P. et al. Constructing and validating of m7G-related genes prognostic signature for hepatocellular carcinoma and immune infiltration: potential biomarkers for predicting the overall survival. J. Gastrointest. Oncol. 13, 3169–3182 (2022).
Liu, Y. et al. Identification of the expression patterns and potential prognostic role of 5-methylcytosine regulators in hepatocellular carcinoma. Front. Cell Dev. Biol. 10, 842220 (2022).
Tong, C., Wang, W. & He, C. m1A methylation modification patterns and metabolic characteristics in hepatocellular carcinoma. BMC Gastroenterol. 22, 93 (2022).
Chang, X. et al. Gene expression profile and prognostic value of m6a RNA methylation regulators in hepatocellular carcinoma. J. Hepatocell. Carcinoma 8, 85–101 (2021).
Sun, Z. et al. Aberrant NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 39, 6906–6919 (2020).
Zhang, X. et al. N-acetyltransferase 10 enhances doxorubicin resistance in human hepatocellular carcinoma cell lines by promoting the epithelial-to-mesenchymal transition. Oxid. Med. Cell Longev. 2019, 7561879 (2019).
Zhao, K. et al. M6A regulator-mediated immune infiltration and methylation modification in hepatocellular carcinoma microenvironment and immunotherapy. Front. Pharmacol. 13, 1052177 (2022).
Li, J. et al. Structural basis of regulated m7G tRNA modification by METTL1-WDR4. Nature 613, 391–397 (2023).
Ruiz-Arroyo, V. M. et al. Structures and mechanisms of tRNA methylation by METTL1-WDR4. Nature 613, 383–390 (2023).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Wang, X. et al. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 (2016).
Schapira, M. Structural chemistry of human RNA methyltransferases. ACS Chem. Biol. 11, 575–582 (2016).
Yankova, E. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021).
Xu, Q. C. et al. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m6A-YTHDF2-dependent manner. Oncogene 41, 1622–1633 (2022).
Moroz-Omori, E. V. et al. METTL3 inhibitors for epitranscriptomic modulation of cellular processes. ChemMedChem 16, 3035–3043 (2021).
Huang, Y. et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell 35, 677–691.e10 (2019).
Liu, Y. et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 33, 1221–1233.e11 (2021).
Feng, P. et al. Inhibition of the m6A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia 36, 2180–2188 (2022).
Cui, Q. et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat. Cancer 2, 932–949 (2021).
Lucas, M. C. et al. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01743-6 (2023).
Sangro, B., Sarobe, P., Hervas-Stubbs, S. & Melero, I. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 18, 525–543 (2021).
Fabbri, L., Chakraborty, A., Robert, C. & Vagner, S. The plasticity of mRNA translation during cancer progression and therapy resistance. Nat. Rev. Cancer 21, 558–577 (2021).
Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010).
Hinnebusch, A. G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 42, 589–611 (2017).
Shi, H., Wei, J. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Acknowledgements
The authors sincerely thank S. Peng, X. Zeng, H. Han, Z. Dai, S. Guo, H. Liu, S. Zheng, H. Guan, Y. Hou and S. Li for their helpful discussions. S.L. is supported by National Key Research and Development Program (2022YFA1105300 and 2022YFE0138700), National Natural Science Foundation of China (82325036 and 81974435), Distinguished Young Scholars grant from Natural Science Foundation of Guangdong (2019B151502011), and Fundamental Research Funds for the Central Universities, Sun Yat-sen University (23ykzy004). M.K. is supported by the Key Program of National Natural Science Foundation of China (82130083), National Science Fund for Distinguished Young Scholars (81825013), and the Key Research and Development Plan of Guangzhou City (202206080016).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Chun-Ming Wong and Puri Fortes for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
NCT05584111: https://clinicaltrials.gov/study/NCT05584111
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, S., Kuang, M. RNA modification-mediated mRNA translation regulation in liver cancer: mechanisms and clinical perspectives. Nat Rev Gastroenterol Hepatol 21, 267–281 (2024). https://doi.org/10.1038/s41575-023-00884-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-023-00884-y