Abstract
Crop yield prediction from UAV images has significant potential in accelerating and revolutionizing crop breeding pipelines. Although convolutional neural networks (CNN) provide easy, accurate and efficient solutions over traditional machine learning models in computer vision applications, a CNN training requires large number of ground truth data, which is often difficult to collect in the agricultural context. The major objective of this study was to develope an end-to-end 3D CNN model for plot-scale soybean yield prediction using multitemporal UAV-based RGB images with approximately 30,000 sample plots. A low-cost UAV-RGB system was utilized and multitemporal images from 13 different experimental fields were collected at Argentina in 2021. Three commonly used 2D CNN architectures (i.e., VGG, ResNet and DenseNet) were transformed into 3D variants to incorporate the temporal data as the third dimension. Additionally, multiple spatiotemporal resolutions were considered as data input and the CNN architectures were trained with different combinations of input shapes. The results reveal that: (a) DenseNet provided the most efficient result (R2 0.69) in terms of accuracy and model complexity, followed by VGG (R2 0.70) and ResNet (R2 0.65); (b) Finer spatiotemporal resolution did not necessarily improve the model performance but increased the model complexity, while the coarser resolution achieved comparable results; and (c) DenseNet showed lower clustering patterns in its prediction maps compared to the other models. This study clearly identifies that multitemporal observation with UAV-based RGB images provides enough information for the 3D CNN architectures to accurately estimate soybean yield non-destructively and efficiently.
Similar content being viewed by others
Introduction
Food insecurity is a major threat to the human civilization given the increasing global population and climate change (Al Hasan et al., 2022; Andreoli et al., 2021). Several studies have estimated that the global food production should be increased by at least 70% by 2050 to meet the demand (Beltran-Pena et al., 2020; Jain et al., 2020; van Dijk et al., 2021). However, the augmentation of agricultural land use and the heightened utilization of natural resources to bolster food production cannot be undertaken without considering the consequential impact on the dynamics of climate change (Fanzo et al., 2021; Wheeler & von Braun, 2013). Therefore, policy makers, breeders, and growers around the world have acknowledged the use of smarter technologies and scientific innovation in crop breeding, which results in hybrids that can produce more yield with fewer natural resources (Edgerton, 2009; Lobell et al., 2009). Accurate and non-destructive estimation of plot-level yield is a significant step in revolutionizing crop breeding operations and understanding the interactions of genetics-management-environment in crop yield (Araus & Cairns, 2014; Song et al., 2021).
Crop yield prediction is a broad topic of interest for crop growers, breeders, and policy makers, simultaneously. While policy makers are concerned with crop yield over larger areas at coarser scale, the growers and breeders are more interested in the variance of crop yield at much finer scales often within the agricultural field (Mcmichael, 1994). Crop growers can be benefited by having an early-stage yield prediction mechanism (van Klompenburg et al., 2020), which can lead to in-field management decisions, such as, precise fertilizer application, irrigation, pest management, etc. Alternatively, crop breeders often use the yield as a metric to evaluate the performance of certain genotypes in development (Hassan et al., 2019). Typically, crop breeding is a multi-year process involving many trials in greenhouses and experimental fields. The trials in experimental fields are more complex as the interactions between different genotypes, management and environmental conditions can affect the yield (Marsh et al., 2021). Therefore, breeders use mechanical harvesters and measure the yield per experimental plot to identify which genotypes or trials perform well in terms of different management and environmental conditions. This is known as the crop breeding pipeline and along with many other plant traits (or phenotypes), yield is one of the most important components of it (Hu et al., 2018). However, measuring yield using harvesters or other destructive means is time consuming and costly in terms of large breeding operations. Therefore, high-throughput, efficient and scalable approaches to crop yield prediction can significantly improve the breeding pipeline and offer much shorter time for seed development (Marsh et al., 2021).
Numerous approaches have been undertaken to conduct crop yield prediction, such as, expert knowledge (Ballot et al., 2018; Papageorgiou et al., 2011, 2013), crop growth models (Basso & Liu, 2019; Huang et al., 2019; Kasampalis et al., 2018), remote sensing methods (Ali et al., 2022; Hara et al., 2021; Khaki et al., 2021; Morales et al., 2023), and hybrid approaches where crop growth models are combined with environmental factors (Gopal & Bhargavi, 2019; Kogan et al., 2013) and remote sensing data (Bai et al., 2019; Zhuo et al., 2019, 2022). While crop growth models are robust and generalized across many different crops and experimental sites, the difficulty often arises during properly parameterizing models for large scale applications and the intrinsic variability of agriculture (Kipkulei et al., 2022).
The plant science community has relied on remote sensing for the past few decades in many different agricultural applications, for instance, estimation of aboveground biomass (Kumar & Mutanga, 2017; Lu et al., 2016;), nitrogen (Knyazikhin et al., 2013; Wang et al., 2017), chlorophyll concentration (Carmona et al., 2015; Guo et al., 2023; Xie et al., 2019), crop yield (Cao et al., 2021; Maimaitijiang et al., 2020; Morales et al., 2023; Sagan et al., 2021), and so on. Since there have been immense developments in spatial, spectral, and temporal resolution in remote sensing products, the high-throughput nature of crop-trait prediction has been receiving new momentum. The principal idea behind using remote sensing techniques in crop trait estimation is that the imageries can capture the spatial and spectral variability of different crops in the field. Such information can then be mapped using linear or non-linear machine learning models, where the independent variables are either the imageries or different features derived from the imageries and the target variable is the measured yield from samples within the experiment.
Many studies have used satellite-level remote sensing and different machine learning models to perform yield prediction for different crops. For example, Schwalbert et al. (2020) utilized Long Short-Term Memory (LSTM) neural networks combined with satellite imagery and weather data to predict soybean yield of Brazil at the municipality level. Peng et al. (2020) used satellite-based Solar-Induced Chlorophyll Fluorescence (SIF) products along with vegetation indices and land surface temperature (LST) from the Moderate Resolution Imaging Spectroradiometer (MODIS) satellite to predict soybean and maize yield for the entire United States. Similarly, regional-scale crop yield prediction has been performed using random forests (Han et al., 2020; Jeong et al., 2016), deep neural networks (Crane-Droesch, 2018), and convolutional neural networks (CNN) based LSTM models (Sun et al., 2019). However, the major challenge with satellite-based remote sensing is that the in-field variation of crop characteristics is not well-captured by the coarser spatial resolution of the satellite data, which does not assist the crop breeders in the breeding pipeline or growers in making management decisions at the within field scale.
Recent advances in unmanned aerial vehicles (UAVs) and sensor technologies have dramatically shifted the capabilities of remote sensing and machine learning in plot-level yield prediction. The higher spatial, spectral, and temporal resolution of UAV sensors offer significant improvements in yield prediction accuracy along with other crop trait estimations. Numerous studies have conducted yield prediction using multispectral (Hassan et al., 2019; Wan et al., 2020), hyperspectral (Feng et al., 2020; Li et al., 2020), or fusion of multi-modal UAV systems (Fei et al., 2022; Maimaitijiang et al., 2020) and achieved consistent results. A major component in these studies was the extraction of hand-crafted features, such as, vegetation indices (i.e., normalized difference vegetation index—NDVI, the wide dynamic range vegetation index—WDRVI), structural (i.e., different height metrics) and textural features (i.e., grey level co-occurrence matrix—GLCM). Extraction of these hand-crafted features requires a priori knowledge of reflectivity and crop biophysical properties. There could be hundreds or thousands of potential features as input for the machine learning model, which can significantly limit the robustness and transferability of the model (Sagan et al., 2021).
A convolutional neural network (CNN) is a specific type of deep learning model, which offers the recognition of spatial and textural patterns within a given sample image. CNN uses convolution and pooling layers to extract meaningful information from complex images and learns to predict a certain target variable. Such capabilities can be leveraged for automatic feature extraction and tasks of hand-crafted feature engineering can be avoided during the model training.
CNN architectures have been proven as highly effective for general computer vision tasks, such as, classification and object detection. The superiority of CNN architectures was first kickstarted with the release of AlexNet (Krizhevsky et al., 2017), followed by VGG (Simonyan & Zisserman, 2015), ResNet (He et al., 2016), DenseNet (Huang et al., 2017), and many more architectures. However, applications of these architectures were mostly confined within general computer vision tasks (Hussain et al., 2019; Nanni et al., 2017) and medical imaging (Singh et al., 2020; Suzuki, 2017). Recently, several studies have employed CNN architectures in direct imagery-based yield prediction from UAV images and have achieved considerable accuracies as well. For example, Yang et al. (2019) concluded that their proposed CNN model provided the most robust rice grain yield forecast compared to other methods. Nevavuori et al., (2019) also found the superiority of CNN models in estimating yield of wheat and malting barley.
While CNN architectures were initially proposed to solve computer vision problems from 2D images, several studies have identified the superiority of 3D CNN for analyzing multitemporal images. In 3D CNN, the images captured from a linear timeseries are stacked together and 3D convolution kernels are used across all three dimensions, i.e., two dimensions for space and the third dimension for time. The principal assumption behind this idea is that 3D convolution can capture both spatial and temporal characteristics from given image samples to predict a target variable. For example, Nevavuori et al. (2020) utilized a CNN-LSTM model and a 3D CNN architecture to estimate crop yield and found that the 3D CNN provided better results compared to the CNN-LSTM model. Similarly, Sagan et al. (2021) employed a 3D version of ResNet architecture for soybean and corn yield prediction, achieved improved performance over traditional machine learning models. An important finding from these studies is being highlighted, namely the superior performance of RGB images in CNN models for yield prediction. Both Yang et al. (2019) and Nevavuori et al. (2019) concluded that RGB images offer more robust CNN training compared to multispectral images. However, many studies have indicated the importance of having multispectral cameras including near-infrared and red-edge bands in yield prediction (Bascon et al., 2022; Shen et al., 2022; Tanabe et al., 2023), Yang et al. (2019) and Nevavuori et al. (2019) highlighted the capability of CNNs to capture important textural and spatial information from the RGB colors in a yield prediction model. Since RGB cameras are more lightweight, low-cost and easy to process, use of RGB images in CNN offers much higher potential in crop breeding and precision agriculture applications.
Although CNN offers automatic feature extraction along with an end-to-end mechanism for crop yield prediction, most of the deep neural networks require massive amounts of ground truth or label data to maintain a robust learning. For typical computer vision and medical imaging tasks, generating ground truth data often involves manual labor and some level of expert knowledge. However, producing ground truth information in terms of agricultural application is a major problem, specifically for regression tasks. For example, a yield prediction problem would require an experimental site with varieties of crop genotypes, management, and environmental conditions. Additionally, the images are required to be collected and plot-level yield information must be measured using harvesters or manual methods. To the best of current knowledge, no studies have been identified that have investigated different CNN architectures for plot-level yield prediction problems for a larger region with a sample size closer to 30,000 plots. Therefore, a knowledge gap exists on the use of specific CNN architecture and optimal spatiotemporal resolution for UAV-based yield prediction.
In this study, end-to-end 3D CNN architectures have been examined for plot-scale soybean yield prediction using multitemporal UAV-based RGB images. Datasets from a soybean breeding pipeline, encompassing a large-scale training sample from different parts of Argentina were incorporated in this study with high variability in genotypes, management, and environmental conditions. To the extent of current awareness, this is the first study to tackle such big data in terms of deep learning and agricultural management within the open-source scientific community. The major objectives of this study are: (a) to determine the best 3D CNN architecture for end-to-end yield prediction, (b) to identify the optimal spatiotemporal resolution for the best prediction accuracy, and (c) to provide an end-to-end solution for big data processing and training in the cloud and offer recommendations for scalability.
Methods
Experimental sites
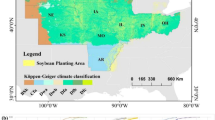
The experimental sites for this study are in Argentina, more specifically in the Province of Buenos Aires, where soybean trials were distributed in four locations. For anonymity of the breeding program, the locations have been codenamed as CON, RAM, ASC, and IND (Fig. 1). ASC and IND are located in the middle, while CON and RAM are in the north of the Province of Buenos Aires. Since all the locations are in the southern hemisphere, the coldest temperatures are found around July, while the warmest temperatures occur around January. All locations have predominant mild weather, which is typical of the Province of Buenos Aires. ASC and IND are 50 km apart from each other; thus, the average temperatures and precipitation are similar. The warmest month is January with historical average temperatures of 23 °C and the coldest month is July with and average temperature of 9 °C. RAM and CON are around 24 km apart; therefore they also share similar average precipitation and temperatures, i.e., the maximum average temperatures in January is 25 °C and the minimum is 11 °C in July. In terms of precipitation, ASC and IND receive an average precipitation of 1043 mm annually, while CON and RAM accumulated 1135 mm of precipitation in the 2021 growing season.
Each location contained several fields, which can be seen in Fig. 1. There were 3 types of trial analyzed in this study. Early (EAL) trials consisted of soybean genotypes in the early stages of the breeding program, where no previous selections have been performed. Intermediate (MID) trials were formed with genotypes in intermediate stages of the breeding program and had gone through previous selection filters. Finally, the advanced (ADV) trials included genotypes with more than one year of evaluation and several selection filters, from which it can be assumed that the genotypes in this trial should have the highest performance consistency throughout locations. All trial fields were planted between October and November of 2020 and harvested between March and May of 2021 (following the different maturity groups of the genotypes in the trials). All trials were managed under no-till schemes and no irrigation, following the typical agricultural practices for the region. The trials also had received similar plot size and distribution. Each plot contained 4 row industry standard dimensions.
Ground truth data collection
The ground truth for this study was yield values measured from each plot in kg.ha−1. The yield value was obtained by harvesting each individual plot with ALMACO Specialized Plot Combines SPC20 (Fig. 2d, ALMACO, Nevada, Iowa, United States). These combines are widely used in the breeding industry. After harvesting each plot, the combines measured humidity and weight of the plot, and stored the information in a local database, where each of these values were associated with its corresponding plot ID. Finally, the plot weight was corrected for humidity by subtracting the weight of the seed due to water content and extrapolated from kg.plot−1 to kg.ha−1. These values were stored in an online database, where each plot was associated with a measured yield value.
UAV data collection
The DJI Phantom 4 Pro UAV (Shenzhen, China) equipped with DJI P4 (Shenzhen, China) multispectral camera (Fig. 2c) was used in this study. The P4 sensor has a total of 6 lenses which captures blue, green, red, red-edge, near-infrared and one dedicated RGB lens. For this study, only the outputs from RGB cameras were used to test different 3D CNN architectures. The flights were performed using DJI GS PRO software (v2.0.17) with a front overlap of 80% and side overlap of 70%. All the flights were performed at a speed of 4.2 ms−1 with images captured every 2 s.
Flight dates were planned based on the soybean phenological cycle, where the focus was to cover all the reproductive stages from R1 to R6 since these periods have been found to be the most important stages for soybean yield prediction. More information about soybean growth stages can be found in (Fehr & Caviness, 1977). First, an initial flight was done at around late vegetative stage (V3-V4) for all locations with the purpose of seeing the plots in a clear way for future plot demarcation. After that, the flights were conducted when the crops were approximately in R1 stage and continued to collect at roughly 2-week intervals (Fig. 3a). However, there were some gaps in the UAV flights due to logistical complexities and bad weather conditions. Approximately nine flights were conducted per site. Figure 3a shows that the gaps between the first and the second flight for ASC and IND are shorter than for CON and RAM. The reason behind this is ASC and IND have genotypes of shorter maturity groups, meaning they reached the reproductive stage earlier than the genotypes in CON and RAM.
The flights were conducted in clear sky conditions. Also, the UAVs were flown near noontime to ensure consistent illumination conditions. Unfortunately, no calibration panel was included for further reflectance calculation, which was one of the major limitations for this study and further explained in the Discussions section. However, the assumption was that since the flights were conducted in clear sky conditions near noontime, the RGB lens DN values would provide normalized information about crop growth. During the flights, several ground control points (GCPs) were placed on the ground which were consistent in between different flights for the same location.
Data processing pipeline
Given the large number of datasets involved in this study, different automatic and semi-automatic measures were adopted in the data processing pipeline. The overall workflow of the pipeline is illustrated in Fig. 4.
Orthomosaic and georeferencing
The orthomosaic images for each field location and date were performed in Pix4D (v4.6.4, Caputo et al., 2023). This process takes individual images captured sequentially and an automated pipeline within the Pix4D software was run to generate RGB orthomosaics. The images from the individual RGB lens of the P4 camera were used which provides an automatically calibrated digital numbers (DN) ranging from 0 to 255. Therefore, any radiometric calibration was not performed to compute reflectance for this lens. Rather the primary objective was to assess the performance of uncalibrated RGB images within CNN models, as many low-cost UAV systems may lack the capability to perform radiometric calibration. Although the UAV had its own GPS and INSS onboard, the resulting orthomosaic still had some errors in terms of accurate locations. Therefore, orthomosaic from a field on one date did not fully align with an orthomosaic from a different date. To fix this problem, the ENVI Image Registration workflow (Xie et al., 2003) was used to automatically connect the consecutive temporal orthomosaics for each field. The Image Registration workflow could identify the GCPs in the fields automatically using its proprietary Registration Engine and used first order polynomial transformation or affine transformation to rectify the georeferenced images. However, this process required manual observations and checking to ensure the quality of the georeferencing outputs. If appropriate overlap was not found within images, a recheck for the tie points was done and adjusted accordingly until the GCPs, and other prominent features aligned with each other.
Plot boundary creation
The plot boundaries from each field were created using both automatic and manual digitizing methods in ArcGIS Pro. First, the starting location for planting was acquired based on how the planter was used to plant soybean seeds in each field. Based on that initial coordinate, a rectangular grid polygon was created based on the known plot width and height information using the Create Fishnet tool in ArcGIS Pro. To check the accuracy of the plot boundaries created by the fishnet tool, a randomized quality checking system was developed involving multiple humans in the process. Such human-in-the-loop mechanism enabled us to remove unnecessary plot polygons or change the shape of the polygon to accurately match the underlying soybean rows. Each plot was uniquely designed with three types of identifiers, i.e., block, range, and column. The three attributes were filled up with semi-automatic fashion in ArcGIS Pro’s automatic numbering tools.
Sample preparation for CNN
The CNN architecture requires the input to be in a multi-dimensional tensor format, where the values in the tensors represent image patches. A fully automatic sample preparation pipeline in Python was generated to efficiently generate the desired sample outputs. First, each plot was used to clip all the temporal images to generate clipped images. Since the orientation of the experimental sites were not the same, the clipped images would have different no data segments for different fields (no data is the black area in Fig. 4d). If such images are used as input for the CNN models, then the CNN might pick up the variation of different orientation of the samples and incorrectly correlate the yield with the no data orientation. Therefore, an automatic rotation algorithm was developed to horizontally rotate each clipped image. First, the clipped images were converted to binary images, where all the no data pixels were assigned “0” and all other value pixels were assigned with “1”. Due to the high contrast of the binary image, the solid boundary lines could be detected using canny edge detectors. Then, the angle of the longest line was calculated and used as the rotation parameter to rotate the images. However, the resulting image then had a significant amount of padding or no data values around the actual pixel values (Fig. 4d), which was reduced using threshold values.
In this study, the effect of spatiotemporal resolution was explored in different CNN architectures while doing yield prediction. Therefore, different sizes of spatial and temporal interpolation for all the image samples was performed. Originally, the 60-m altitude UAV flight resulted in around 3.4 cm ground sampling distance (GSD) resolution. Spatial interpolation was done using the nearest neighbor method and it was necessary because the image samples had ununiform spatial dimensions after clipping, rotation, and padding removal. Table 1 shows the different sizes of spatial grid size considered for experiment in this study. Alternatively, only 9 temporal dimensions were available as the depth of each image. The architectures used in this study required more than 9 temporal dimensions to run all the convolution and pooling layers within the architecture. Therefore, each pixel and its corresponding 9-pixel values were considered as independent timeseries (from 9 temporal images), and different sizes of values, i.e., 32, 48, 64, 80, were interpolated. The linear interpolation was performed by considering the band values for each pixel as a timeseries and populating the intermediate values by considering the number of days between each image acquisition. Table 1 shows all the considerations of different spatial and temporal sizes for experiments within the study. The number 32 was simply picked as the base number of sizes and incremented with 16 for different combinations of input configuration.
CNN architectures
Numerous studies in the field of crop yield prediction have employed CNNs, with many constructing bespoke architectures by iteratively adding layers with varying input dimensions, kernel shapes, and filter configurations (Nevavuori et al., 2019, 2020; Yang et al., 2019). However, in the realm of computer vision, where millions of training samples are readily available in standardized datasets, CNNs have evolved into various well-established architectures like VGG, ResNet, DenseNet, among others. While availability of such architectures would ensure scalability, robustness and transferability in a crop breeding pipeline, evaluating these architectures for crop phenotyping or yield prediction becomes challenging due to the scarcity of large-scale samples in this specialized field. Therefore, a substantial dataset of approximately 30,000 instances with accompanying ground truth yield data was leveraged to analyze three fundamental and widely recognized architectural types (i.e., VGG, ResNet, and DenseNet) to discern the most accurate and efficient performer, regardless of input spatiotemporal dimensions.
Neural networks have shown improved performances over many different classifications and regression problems. However, deep neural networks often do not share parameters well in terms of 2D images. Convolution is a mathematical operation that allows the merging of spatial information in a 2D space. Therefore, 2D convolutions are the most popular operations in many CNN architectures where they extract useful information from a complex 2D space.
The 2D input image can have multiple channels, which is 3 in this study (i.e., red, green and blue), and multiple convolution kernels with \(k\times k\) size can be acted as a sliding window over the image to perform the following mathematical operation:
where, \(t\) and \(h\) represent the \(t\)-th sliding window and \(h\)-th convolution kernel, respectively; \({k}_{h}\) and \({k}_{w}\) are the spatial kernel sizes of \(h\) height and \(w\) width; \({x}^{(t)}\) and \({z}^{(t)}\) are the \(t\)-th input and \(t\)-th output, and \({y}^{(h)}\) and \({b}^{(h)}\) denote the \(h\)-th kernel matrix and its bias, respectively. The output from a single channel convolution is a 2D feature map, where the shape is expressed as \((h-{k}_{h}+1)\times \left(w-{k}_{w}+1\right)\) map.
Similarly, 3D convolution is an intuitive extension of 2D convolutions that can consider an extra dimension for more information extraction. In this case, the 3rd dimension is the temporal dimension, which represents sequential growth patterns of soybean in the field. The 3D convolution operation for a 3D input matrix can be expressed as Eq. 2.
where, \({k}_{d}\) is the depth of kernels representing the temporal information. In 3D convolution, a 3D kernel slides around the 3D input matrix and summarizes the information content across all the channels. The resulting output from 3D convolution is also a 3D matrix, where the shape is \((h-{k}_{h}+1)\times \left(w-{k}_{w}+1\right)\times \left(d-{k}_{d}+1\right)\) map.
In this study, the three most used CNN architecture families (i.e., VGG, ResNet, and DenseNet) were considered as the basis of the experiment. However, the CNN architectures were initially proposed to handle 2D images for classification problem, whereas the dimensionality of these architectures were extended to match the input–output shape of a 3D matrix. In the following section, the major configuration of these architectures and their 3D representation for yield prediction problem are discussed.
VGG
The family of VGG (Visual Geometry Group) architectures was first proposed by Simonyan and Zisserman (2015) as an improvement to AlexNet (Krizhevsky et al., 2017) and ZFNet (Zeiler & Fergus, 2014). The VGG family was proposed with several levels of network depth consisting of 11, 13, 16, and 19 layers. The network consists of a number of repeating building blocks, initialized with a stack of convolution layers up to four times and adjacent dimension reduction by max pooling. The architecture used \(3\times 3\) kernel size exclusively in the convolutional layers. However, the kernel size was extended from \(3\times 3\) to \(3\times 3\times 3\) to match the 3D input shape of the yield prediction problem. After each convolution, the ReLU (Rectified Linear Unit) was used as the activation function, which calculates \(f\left(x\right)=\underset{}{{\text{max}}}(0,x)\) on the resulting feature maps (Nair & Hinton, 2010). The VGG-13 was chosen as one of the candidates from the VGG family architecture where 13 layers of convolution and pooling operations are performed (Fig. 5a). After the feature map generation from the convolution and pooling layers, a global average pooling was performed from the 3D feature map and two dense layers were used followed by an output layer with one neuron to predict yield.
The schematic diagram of the three 3D CNN architectures, i.e., VGG-13 (a), ResNet-50 (b), and DenseNet-169 (c). VGG-13 includes several convolution and pooling layers as the building block of the architecture. The ResNet-50 has two types of residual blocks building the entire network. Alternatively, DenseNet-169 consists of dense and transition blocks as the main backbone
ResNet
The ResNet family was first introduced in 2015 by He et al., (2016) to account for the vanishing gradient problem of the CNN architectures. While building very deeper networks, the computer vision community identified the vanishing gradients or exploding gradients, where the value of the product of derivative decreases until the partial derivative of the loss function approaches a value close to zero (Ide & Kurita, 2017; Su et al., 2018; Yang et al., 2022a, 2022b). This phenomenon often leads to poor learning or no learning at all at the deeper layers resulting in decreased performance. ResNet architectures avoid the degradation of deeper networks and support the optimization with backpropagation by reformulating the function \(H(x,{W}_{i})\), which is the convolutional operations of the main trunk. The function acts as a residual function \(H\left( {x,W_{i} } \right)\, = \,F\left( {x,W_{i} } \right) + x\), where \(F\left(x,{W}_{i}\right)\) is the residue to approximate and \(x\) is the input before the convolutional operation. Approximation of the residue is easier as \(x\) is known due to the residual connection. Moreover, an identity function is learned from \(x\) since the residue of convolutional operations in final part of the network is zero. In terms of the backpropagation point of view, the transportation of larger gradients to convolution operations is enabled and optimization of those layers is confirmed. Therefore, residual connections offer better performance in deeper networks compared to shallower networks (Xie et al., 2017; Yang et al., 2022a, 2022b).
If a network is considered with \(L\) layers, where each layer performs a non-linear transformation \({H}_{L}\), the output of the \({L}_{th}\) layer is denoted as \({X}_{L}\) and the input image is represented as \({x}_{0}\), then the skip connection can be represented as Eq. 3.
ResNet was proposed with multiple depths, for instance, 18, 34, 50, 101, and 152 layers. However, the ResNet-50 was considered as the candidate for yield prediction model (Fig. 5b). Similar to the VGG-13 network, all the 2D convolution and pooling layers were transformed into 3D shape to accommodate the spatiotemporal input data. The final output layer of the network also consists of one neuron to directly predict yield. Figure 5b shows the overall architecture of ResNet-50 along with the schematic diagram of residual blocks.
DenseNet
DenseNet is a variant of residual network that was first proposed in 2017 by Huang et al., (2017). Whereas in ResNet, identify mapping is performed to promote the gradient propagation using element-wise addition, DenseNet obtains additional inputs from all preceding layers and passes on its own feature maps to the subsequent layers (Fig. 5c). The DenseNet network is divided into multiple densely connected dense blocks to facilitate both down-sampling and feature concatenation. Such division of the network into dense blocks allows the concatenation of feature maps from different sizes. Moreover, transition layers are also used in between the dense layer which consists of a batch normalization layer, \(1\times 1\times 1\) convolution followed by a \(2\times 2\times 2\) average pooling layer. Because of the use of concatenation between dense blocks, each layer learns a collective knowledge from all preceding layers resulting in a thinner and compact network. Therefore, DenseNet require fewer model parameters to train and provide higher computation efficiency (Chen et al., 2021).
The difference between a skip connection of a ResNet and the denseonnectivevity in DenseNet architecture can be seen in Eq. 4 in terms of the variables set for Eq. 3.
where, \([{X}_{0},{X}_{1},\dots ,{X}_{l-1}]\) represents the concatenation of the feature maps produced by \(\mathrm{0,1},\dots ,{L}_{th}\) layers.
Different depths of DenseNet architectures were originally proposed, i.e., 121, 169, 201, and 264 layers. The DenseNet-169 network was chosen for this study. Similar to the VGG and ResNet architecture, the convolution and pooling layers were transformed into 3D format and the final output layer was considered with one neuron to perform yield prediction.
Model training
The models were trained using cloud solutions provided by Amazon Web Services (AWS). The AWS Sagemaker (v2.140.1) platform was used to train and evaluate the performance of the three architectures for different input configurations shown in Table 1. The automatic sample preparation pipeline was utilized to generate different sizes of input configurations and uploaded to AWS S3 bucket. During the model training, the input data was directly fed from the corresponding S3 bucket. The TensorFlow API (v2.6.0) was used for training the models in AWS Sagemaker.
The training configuration for each architecture and input shape was kept similar so the experiments return a fare comparative result. First, the sample dataset for each input configuration was divided into three different sets, i.e., training, validation and test set using a randomized 50%, 20% and 30% split, respectively. The sample image values ranged from 0 to 255 in a 32-bit data structure. Therefore, each sample was divided by 255 to scale the images from 0 to 1. Since the problem in this study is a regression problem, mean squared error was considered as the loss function for all the model training. The Adam algorithm (Kingma & Ba, 2015) was utilized to optimize for the loss function, which has proved to be highly efficient in most of the advanced deep learning problems.
A standardized set of hyperparameters were considered during the model training. The batch size was kept as 32 for all the training. The learning rate was initially set at 0.0001 and slowly reduced with the increasing iterations. The total number of iterations were set at 200, but an early stopping criterion was used to automatically stop the model training if the validation set loss does not improve. During the model training, both training and validation loss were retained for further evaluation.
Model evaluation
The performance of each model was evaluated based on three common regression metrics, i.e., coefficient of determination (\({R}^{2}\)), root mean squared error (\(RMSE\)), and normalized root mean squared error (\(NRMSE\)), which are expressed in following equations:
where, \(i=\mathrm{1,2},3,\dots \dots ..,n\) is the test sample, \({\widehat{y}}_{i}\) and \({y}_{i}\) represent the predicted and measured yield values, respectively, and \(\overline{y }\) is the average of each measured values.
The Global Moran’s I statistic (Anselin, 1995; Harries, 2006) of the regression residuals was also calculated for each plot, field, and model. First, each trained model was applied to all the plots for each field and then the residuals were analyzed with Global Moran’s I. Understanding the spatial heterogeneity and dependency of crops over the experimental fields is an important consideration for yield prediction as crop breeding often suffer from intrinsic spatial dependence (Haghighattalab et al., 2017). Therefore, many studies have accounted for the effect of spatial autocorrelation in regression-based estimations (Anselin et al., 2004; Ghulam et al., 2015; Maimaitijiang et al., 2015; Peralta et al., 2016). The Global Moran’s I measure the spatial autocorrelation or dependence based on both feature locations and values simultaneously. It simply explains if a given set of values is clustered, dispersed, or random in terms of a given spatial scale by resulting in a Moran’s Index and associated p-value for statistical significance. A positive Moran’s Index indicates that the spatial pattern is clustered, whereas a negative index represents dispersed pattern in a given space. However, if the p-value associated with the Moran’s Index is not statistically significant, then it can be inferred that there exists a complete spatial randomness in the model residuals.
Results
Descriptive statistics of yield
The descriptive statistics for the target variable, yield, are shown in Fig. 6 and Table 2. When all the datasets are considered (32,272 samples), the distribution of yield shows a normal distribution (Fig. 6a) with 3610 kg ha−1 as the mean and 663 kg ha−1 as standard deviation (Table 2). In terms of different trials, the values of yield also show a normal distribution (Fig. 6b), where the number of samples in MID trials are the highest followed by EAL and ADV (Fig. 2b and Table 2). As per the previous discussion, EAL trial includes soybean genotypes that were in a very early stage of the breeding program, which might result in an extreme variation of its yield values. Such phenomenon can be observed in Table 1 by looking at the range values. The range of the EAL trials (6225 samples) is significantly higher than the MID (4819 samples) and the ADV (4621 samples). The interquartile range also follows a similar pattern in the yield values. However, the distribution of yield values over different trials and fields shows a variety of patterns as well. Figure 6c visualizes the distributions as multiple boxplots for 13 different experimental fields. Each field may or may not contain multiple trials. For example, the ASC-A1 and CON-C4 fields only had EAL trials, whereas IND-I2, RAM-R5, and RAM-R4 had all three trials. All other fields contained at least a combination of MID and ADV trials. Figure 6c clearly shows that the range (total spread of the lines) and the interquartile range (the height of each box) were comparatively higher for the ERL trials, which indicates the higher variability of yield in earlier stages of corn breeding. The median yield value in ERL trials (the middle line in the boxplots) also tends to be higher than the median of corresponding trials. Although the amount of ERL samples is the second highest compared to MID and ADV, only 6 fields among 13 had ERL trials.
Descriptive statistics of the yield, where a shows the distribution of the entire dataset, b visualizes the distribution of yield over three trials in the breeding program (i.e., ERL, MID, and ADV), and c shows the individual distribution of yield over both trial and fields. In boxplots: whiskers show maximum and minimum values, the box shows the interquartile range, the middle line is the median
Prediction results
The test set model evaluation metrics are shown in Table 3 and the R2 of different architectures is visually compared in Fig. 7. Overall, the VGG-13 and DenseNet-169 both outperformed the ResNet-50 in terms of model accuracy. The highest R2 (0.70) and the lowest NRMSE (10.08%) were achieved by VGG with the input shape of \(80\times 80\times 32\) (Table 3). However, the VGG also performed very similarly with the input shape of \(48\times 48\times 32\) (R2 = 0.70 and NRMSE = 10.15%), which indicates the usefulness of even smaller spatial resolution in imagery-based yield prediction. The second-best result was achieved by DenseNet with the input shape of \(80\times 80\times 48\) (R2 = 0.69 and NRMSE = 10.33%), followed by \(48\times 48\times 32\) (R2 = 0.69 and NRMSE = 10.44%) with a slight difference in the NRMSE. The overall performance of ResNet was comparatively poorer than VGG and DenseNet. The best ResNet performance was achieved with \(64\times 64\times 64\) input shape (R2 = 0.65 and NRMSE = 11.05%).
Coefficient of determination (R2) of test set data for VGG, ResNet, and DenseNet architectures with different input shapes. The x-axis shows the varying input shapes, y-axis in the left side shows the R2 and the y-axis in the right side shows the number of trainable parameters in millions. The dashed lines correspond to the right-side y-axis representing the number of parameters
In terms of different trials, the trained CNN architectures performed relatively well in predicting unseen samples from ADV compared to MID and ERL samples (Fig. 8). For example, VGG resulted in an R2 of 0.71 and NRMSE of 9.63% in ADV, whereas it slightly underperformed for MID (R2 = 0.67 and NRMSE = 10.00%) and ERL (R2 = 0.67 and NRMSE = 10.32%). A similar pattern was also observed for DenseNet (Fig. 8b, 10e, and h) and ResNet (Fig. 8c, f, and i). Since ERL trials included lots of earlier candidates of soybean genotypes in the breeding operations, the lower performance in ERL trials is expected. However, most of the models consistently provided the best performance in ADV trials, which included the most advanced soybean lines in the breeding pipeline. It should be noted that crop breeders are more interested in accurate yield prediction of the commercially successful trials rather than preliminary trials, which was achieved by the 3D CNN architectures for this particular experiment (Fig. 8g–i).
Figure 8 also highlights that the inconsistencies between the measured and predicted yield are concentrated at the lower yield samples compared to the higher yield samples (marked with blue dashed circles in Fig. 8). This phenomenon was further quantified by dividing the test set prediction results into 4 quartiles (i.e., 0–25th percentile, 25–50th percentile, 50–75th percentile, and 75–100th percentile) of the measured yield range. Figure 9 shows the plotting of absolute error over different quartiles of measured yield and corresponding NRMSE value. It is evident that the NRMSE value is consistently the highest in 1st quartile, followed by the 4th quartile, which indicates that model could successfully explain the yield variability in the middle and extreme quartiles, whereas most of the errors are concentrated in the lower performing soybean plots in terms of yield.
Efficient CNN architecture
The results from this study clearly indicate the efficacy of 3D CNN architectures in understanding the spatiotemporal characteristics of soybean growth and accurately predicting soybean yield. Both VGG and DenseNet outperform the result of ResNet in overall yield prediction. Sagan et al., (2021) reported that the 3D version of ResNet provided the best performance over hand-crafted feature-based yield predictions from traditional machine learning algorithms (i.e., SVR, RFR, and PLSR) while using WorldView and PlanetScope satellite information. However, this study shows that a typical vintage architecture (i.e., VGG) and a more simplified residual network (i.e., DenseNet) can outperform ResNet. It is noteworthy to highlight that ResNet is a more costly architecture in terms of the model parameters compared to VGG and DenseNet. Figure 7 shows three lines representing the number of trainable parameters for different architectures and ResNet has a significantly higher number of model parameters compared to others. For example, the best model from ResNet was found for the input of \(64\times 64\times 64\) with around 130 million parameters, whereas the best model from VGG and DenseNet had approximately 53 million and 42 million parameters, respectively. As per previous discussion, ResNet uses residual skip connections and additions that results in a higher number of trainable parameters. Alternatively, DenseNet, which is a more efficient version of residual network, uses concatenation in its dense blocks and results in a smaller number of parameters regardless of the input shape. CNN architectures that are lightweight in that parameters provide faster training and prediction inference from large amount of data (Hoeser & Kuenzer, 2020). Therefore, it can be concluded that DenseNet provided the most efficient performance in yield prediction as the evaluation metrics were consistent yet with a smaller number of model parameters and providing much faster training and inference speed.
Spatial analysis of yield prediction
The spatial pattern of predicted yield for each plots in each field was performed by using the Global Moran’s I statistic. Figure 10 shows the prediction map generated from the best performing DenseNet model in one field from each location. Similar results was generated for all the fields and models within this study and the Global Moran’s I analysis was performed for all the models and illustrated in Fig. 11.
Spatial autocorrelation result of the prediction residuals for all samples in different fields and models. The positive Moran’s Index indicates spatial clustering, whereas the negative value shows a dispersed spatial pattern. The bars with an asterix (*) indicate the Morna’s I index of statistical significance at 95% confidence interval (\(p<0.05\))
The Global Moran’s I analysis resulted in some statistically insignificant result considering 95% confidence interval (\(p<0.05\)). However, some of the fields returned statistically significant results and in most of the cases, DenseNet showed comparatively lower clustering pattern than the other models. The clustering could be resulted due to the planting of soybean plots in similar trials within blocks. However, in terms of spatial autocorrelation, DenseNet tends to outperform the other models in providing less clustered residuals of yield over space.
Discussions
Effectiveness of RGB images in yield prediction
The contribution of spectral information through different vegetation indices (VIs), structural, and textural features have been well documented in different remote sensing literature for crop trait estimation (Ballester et al., 2017; Zhou et al., 2017). Many studies have utilized CNNs with UAV-based multispectral sensor with near-infrared and red-edge bands for yield prediction. For example, Li et al. (2023) recommended CNN as the best performing model when compared with three statistical machine learning models (i.e., random forest, gradient boosting machine, K-nearest neighbor) for winter wheat yield prediction using 16 yield-sensitive Vis. Similarly, Tanabe et al. (2023) and Bellis et al. (2022) found the superiority of CNN in winter wheat and rice yield prediction by using several multispectral-derived Vis. In most of these studies, near-infrared band-based Vis seemed to be highly informative for yield prediction performance.
However, when direct images are concerned in predictive models instead of hand-crafted VIs, several studies have found that RGB images provide better and robust predictive models over VI-based models (Nevavuori et al., 2019; Yang et al., 2019). The Canopy Height Model (CHM) and Vegetation Fraction (VF) information derived from RGB images were often found to be highly informative over VI-based spectral features in understanding plant characteristics (Carly et al., 2017; Maimaitijiang et al., 2017; Rischbeck et al., 2016; Wallace, 2013). The probable reason behind this could be that direct imagery-based CNN models have the capabilities of understanding the spatial variation of canopy architectures in a given plot, and therefore, accurate mapping of yield per plot is possible in an efficient manner. Additionally, the inclusion of multi-temporal observation enables the 3D CNN architectures to understand the temporal growth pattern of crops over space, whereas a tabular feature-based modeling would not consider the spatial and temporal dependency of features. Considering these scenarios, the RGB image-based yield prediction was considered but more focus was given on the specific type of CNN architecture and spatiotemporal resolution for this task. It is possible that the inclusion of near-infrared bands might improve the model performance, but it increases the complexity of expensive camera system, higher battery consumption of UAVs, and decreases the amount of training data in the process. Alternatively, use of RGB-equipped UAV provides the advantages of simple flight operations, easier image calibration and post-processing tasks in the modeling pipeline.
Effect of spatiotemporal resolution
Although the utilization of custom CNN architectures was previously examined in different studies, it was unclear whether there is any significant effect of spatiotemporal resolution in model performance. Based on the flight altitude of the UAV, an approximated 3.4 cm spatial resolution was achieved for the orthomosaic. However, the spatial resolution was increased to three different levels (Table 1) and tested these with different CNN architectures. The results suggest that even with coarser spatial resolution, the architectures could achieve comparable performance in yield prediction. While different literature reported the use of 3–4 cm spatial resolution for each sample plot in the CNN architectures (Nevavuori et al., 2019, 2020; Yang et al., 2019), the VGG and DenseNet architecture could achieve comparable performance using \(48\times 48\) as the spatial grid size resembling approximately 8.93 cm ground resolution.
In terms of temporal resolution, the temporal dimension was increased from 9 to 32, 48, 64, and 80 by performing linear interpolation at the temporal scale. This was done to accommodate the deeper 3D CNN architectures addressed in this study. If only 9 dimensions were used as the temporal resolution, then the 3D versions of VGG, ResNet and DenseNet could not be trained as negative dimension would be caused from fewer dimensions. The temporal interpolation enabled the architectures to learn about the smooth transition of soybean growth over time. The importance of enabling temporal observation in CNN and deep learning has also been reported in multiple studies mainly for crop type classification and yield prediction (Ghamisi et al., 2019; Hauglin & Orka, 2016; Ji et al., 2018; Nevavuori et al., 2020; Sun et al., 2020). However, it was not observed that increasing the temporal dimension would necessarily improve the model performance as the highest performance from both VGG and DenseNet was achieved with only 32 temporal dimensions. The recommendation arises that a 2-week interval UAV flight can accurately explain the growth variability of soybean in a 3D CNN-based model training.
Integration of end-to-end modeling in crop breeding
Improving crop yield is a fundamental goal in achieving food security and sustainable agricultural management. Crop breeding offers significant advancement of genetic technologies and high-throughput phenotyping (HTTP) to achieve such yield goals with fewer natural resources (Araus & Cairns, 2014). However, it still takes years to breed a successful genotype given the high variability of climate change and other natural hazards. An end-to-end yield prediction pipeline involving low-cost RGB cameras and UAVs can offer significant breakthroughs in the bottleneck of crop breeding (Shakoor et al., 2017; Song et al., 2021). The findings from this study can be easily integrated into the current crop breeding pipeline and reduce the time required for typical breeding. It was also found that a 3D version of DenseNet could accurately explain the variability of soybean yield with the use of RGB images, which is easier to collect and process. Although the analysis was done for the 2020–2021 growing season involving four major locations for soybean crops, the methodology can be easily extended to other study areas and crops in the future. Additionally, the concept of transfer learning can be applied to this scenario (Tan et al., 2018; Zhuang et al., 2021), where a model is trained from a large number of datasets and later used to retrain with a smaller batch of datasets. Moreover, the use of AWS cloud infrastructure in training the models offers significant scalability in the future as well. A prediction model can be easily hosted as an endpoint in the cloud and predict future yield from newer sets of UAV-based data.
Limitations and future steps
In this study, a UAV equipped with a multispectral sensor that can collect blue, green, red, red-edge, and near-infrared information simultaneously. However, only the RGB information was used instead of the multispectral data because of incorrect radiometric calibration issues. The DJI P4 sensor offers a sunshine sensor that collects simultaneous sun irradiance (in W m−2) values for each scene capture. The camera documentation suggests that the sunshine sensor is accurate enough to perform the radiometric calibration without any use of a reference panel on the ground. However, literature suggests that use of a factory-tested calibration panel during the UAV data collection procedure offers accurate calculation of reflectance values for analysis (Cao et al., 2019; Iqbal et al., 2018). After processing the orthomosaic and radiometric calibration in Pix4D, incorrect reflectance values were observed from the multispectral bands. For instance, the value of the blue band was found to be significantly higher than the corresponding green or red band, which is theoretically not possible in terms of healthy vegetation. Therefore, any yield prediction modeling for the multispectral images was not performed. However, the use of a factory-tested calibration panel during data collection is being considered at this moment, and in future a comparative assessment of will be done for correctly calibrated multispectral images in yield prediction.
Another limitation for this study was using a dataset from a single growing season. If there were multiple growing seasons with data for similar genotypes and experimental sites, then the training could be done on one or two seasons of data, while the performance of the models could be validated using data for a different season. However, the major objective for this work was to evaluate the performance of some state-of-the-art CNN architectures in yield prediction and assess which end-to-end system had the most efficient performance. The comparison was done for all the architectures using the same train-test splitting strategy, which made it possible to make robust inference about the model performance. In future, more growing season data will be accumulated and will be used with the most efficient model from this study (i.e., DenseNet). This will directly enable a more robust splitting strategy.
The major reason behind not training any hand-crafted feature-based modeling in this study was the lack of calibrated reflectance values from the RGB lens. Typically, in a tabular feature space, many different vegetation indices are calculated first and then different statistical machine learning models, such as, random forest, support vector machines, or extreme gradient boosting algorithms are used to train a model. Since the dataset from this study only had scaled 8-bit information as DN, calculation of vegetation indices would be unrealistic. Furthermore, the investigation aimed to explore the potential utility of CNNs within a crop breeding pipeline. While it was evident that CNNs can learn to estimate yield, the models can be further enhanced by implementing transfer learning with datasets from other crops, growing seasons, and locations. On the other hand, statistical machine learning algorithms have much shallower architecture, which limits the practicality of using transfer learning.
Conclusions
In conclusion, this study presents a comprehensive end-to-end 3D CNN framework for plot-level soybean yield estimation utilizing multitemporal Unmanned Aerial Vehicle (UAV)-based RGB imagery. To the best of current knowledge, this is the first study of its kind, incorporating approximately 30,000 samples across various fields and multiple CNN architectures for the purpose of soybean yield prediction. Among the key discoveries, DenseNet emerged as the most effective architecture in terms of both accuracy and model complexity for yield prediction, with VGG and ResNet following suit. Although VGG exhibited marginal improvements in evaluation metrics compared to DenseNet, it proved more resource-intensive in terms of training and inference. Furthermore, it was observed that higher spatiotemporal resolution of input samples did not enhance model performance, but instead increased the complexity for VGG and ResNet. A coarser spatiotemporal resolution of \(48\times 48\times 32\) achieved comparable performance in yield prediction to finer resolutions. In addition, DenseNet displayed reduced clustering patterns in its prediction maps relative to other models. Overall, the study demonstrated that multitemporal observations employing UAV-based RGB images supplied sufficient information for the 3D CNN architectures to accurately and efficiently estimate soybean yields in a non-destructive manner.
In summary, the results and the development of the end-to-end 3D CNN pipeline underscore the potential for employing cost-effective RGB-based UAV systems in soybean yield prediction, with applications in crop breeding operations and precision agriculture. Nevertheless, it is imperative to evaluate the model’s efficacy across various test sites, crops, and temporal scales. Additionally, incorporating secondary datasets (e.g., weather station data, soil sensor information) as separate branches in the prediction model, in conjunction with the 3D CNN, could further enhance its performance. Consequently, the insights gathered from this study offer valuable guidance on selecting appropriate architectures, determining optimal input configurations, and managing vast amounts of data.
References
Al Hasan, S. M., Saulam, J., Mikami, F., Kanda, K., Ngatu, N. R., Yokoi, H., & Hirao, T. (2022). Trends in per capita food and protein availability at the national level of the Southeast Asian Countries: An analysis of the FA’’s food balance sheet data from 1961 to 2018. Nutrients. https://doi.org/10.3390/nu14030603
Ali, A. M., Abouelghar, M., Belal, A. A., Saleh, N., Yones, M., Selim, A. I., Amin, M. E. S., Elwesemy, A., Kucher, D. E., Maginan, S., & Savin, I. (2022). Crop yield prediction using multi sensors remote sensing. The Egyptian Journal of Remote Sensing and Space Science, 25(3), 711–716. https://doi.org/10.1016/j.ejrs.2022.04.006
Andreoli, V., Bagliani, M., Corsi, A., & Frontuto, V. (2021). Drivers of protein consumption: A cross-country analysis. Sustainability. https://doi.org/10.3390/su13137399
Anselin, L. (1995). Local indicators of spatial association—LISA. Geographical Analysis, 27(2), 93–115. https://doi.org/10.1111/j.1538-4632.1995.tb00338.x
Anselin, L., Bongiovanni, R., & Lowenberg-DeBoer, J. (2004). A spatial econometric approach to the economics of site-specific nitrogen management in corn production. American Journal of Agricultural Economics, 86(3), 675–687. https://doi.org/10.1111/j.0002-9092.2004.00610.x
Araus, J. L., & Cairns, J. E. (2014). Field high-throughput phenotyping: The new crop breeding frontier. Trends in Plant Science, 19(1), 52–61. https://doi.org/10.1016/j.tplants.2013.09.008
Bai, T. C., Wang, S. G., Meng, W. B., Zhang, N. N., Wang, T., Chen, Y. Q., & Mercatoris, B. (2019). Assimilation of remotely-sensed LAI into WOFOST model with the SUBPLEX algorithm for improving the field-scale jujube yield forecasts. Remote Sensing. https://doi.org/10.3390/rs11161945
Ballester, C., Hornbuckle, J., Brinkhoff, J., Smith, J., & Quayle, W. (2017). Assessment of in-season cotton nitrogen status and lint yield prediction from unmanned aerial system imagery. Remote Sensing. https://doi.org/10.3390/rs9111149
Ballot, R., Loyce, C., Jeuffroy, M. H., Ronceux, A., Gombert, J., Lesur-Dumoulin, C., & Guichard, L. (2018). First cropping system model based on expert-knowledge parameterization. Agronomy for Sustainable Development. https://doi.org/10.1007/s13593-018-0512-8
Bascon, M. V., Nakata, T., Shibata, S., Takata, I., Kobayashi, N., Kato, Y., Inoue, S., Doi, K., Murase, J., & Nishiuchi, S. (2022). Estimating yield-related traits using UAV-derived multispectral images to improve rice grain yield prediction. Agriculture. https://doi.org/10.3390/agriculture12081141
Basso, B., & Liu, L. (2019). Seasonal crop yield forecast: Methods, applications, and accuracies. Advances in Agronomy, 154(154), 201–255. https://doi.org/10.1016/bs.agron.2018.11.002
Bellis, E. S., Hasehm, A. A., Causey, J. L., Runkle, B. R. K., Moreno-Garcia, B., Burns, B. W., Green, V. S., Bursham, T. N., Reba, M. L., & Huang, X. (2022). Detecting intra-field variation in rice yield with unmanned aerial vehicle imagery and deep learning. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2022.716506
Beltran-Pena, A., Rosa, L., & D’Odorico, P. (2020). Global food self-sufficiency in the 21st century under sustainable intensification of agriculture. Environmental Research Letters. https://doi.org/10.1088/1748-9326/ab9388
Cao, J., Zhang, Z., Luo, Y. C., Zhang, L. L., Zhang, J., Li, Z. Y., & Tao, F. L. (2021). Wheat yield predictions at a county and field scale with deep learning, machine learning, and google earth engine. European Journal of Agronomy. https://doi.org/10.1016/j.eja.2020.126204
Cao, S., Danielson, B., Clare, S., Koenig, S., Campos-Vargas, C., & Sanchez-Azofeifa, A. (2019). Radiometric calibration assessments for UAS-borne multispectral cameras: Laboratory and field protocols. ISPRS Journal of Photogrammetry and Remote Sensing, 149, 132–145. https://doi.org/10.1016/j.isprsjprs.2019.01.016
Caputo, T., Sessa, E. B., Marotta, E., Caputo, A., Belviso, P., Avvisati, G., Peluso, R., & Carandente, A. (2023). Estimation of the uncertainties introduced in thermal map mosaic: A case of study with PIX4D mapper software. Remote Sensing, 15(18), 4385. https://doi.org/10.3390/rs15184385
Carly, S., Michael, J. S., Norman, E., Michael, B., Murilo, M. M., & Tianxing, C. (2017). Unmanned aircraft system-derived crop height and normalized difference vegetation index metrics for sorghum yield and aphid stress assessment. Journal of Applied Remote Sensing, 11(2), 026035. https://doi.org/10.1117/1.JRS.11.026035
Carmona, F., Rivas, R., & Fonnegra, D. C. (2015). Vegetation Index to estimate chlorophyll content from multispectral remote sensing data. European Journal of Remote Sensing, 48, 319–326. https://doi.org/10.5721/EuJRS20154818
Chen, B., Zhao, T., Liu, J., & Lin, L. (2021). Multipath feature recalibration DenseNet for image classification. International Journal of Machine Learning and Cybernetics, 12(3), 651–660. https://doi.org/10.1007/s13042-020-01194-4
Crane-Droesch, A. (2018). Machine learning methods for crop yield prediction and climate change impact assessment in agriculture. Environmental Research Letters. https://doi.org/10.1088/1748-9326/aae159
Edgerton, M. D. (2009). Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiology, 149(1), 7–13. https://doi.org/10.1104/pp.108.130195
Fanzo, J., Bellows, A. L., Spiker, M. L., Thorne-Lyman, A. L., & Bloem, M. W. (2021). The importance of food systems and the environment for nutrition. American Journal of Clinical Nutrition, 113(1), 7–16. https://doi.org/10.1093/ajcn/nqaa313
Fehr, W., & C. Caviness. (1977). Stages of Soybean Development. Iowa State University (Ames, Iowa: Iowa State University). https://dr.lib.iastate.edu/handle/20.500.12876/90239.
Fei, S. P., Hassan, M. A., Xiao, Y. G., Su, X., Chen, Z., Cheng, Q., Duan, F. Y., Chen, R. Q., & Ma, Y. T. (2022). UAV-based multi-sensor data fusion and machine learning algorithm for yield prediction in wheat. Precision Agriculture. https://doi.org/10.1007/s11119-022-09938-8
Feng, L. W., Zhang, Z., Ma, Y. C., Du, Q. Y., Williams, P., Drewry, J., & Luck, B. (2020). Alfalfa yield prediction using UAV-based hyperspectral imagery and ensemble learning. Remote Sensing. https://doi.org/10.3390/rs12122028
Ghamisi, P., Rasti, B., Yokoya, N., Wang, Q. M., Hofle, B., Bruzzone, L., Bovolo, F., Chi, M. M., Anders, K., Gloaguen, R., Atkinson, P. M., & Benediktsson, J. A. (2019). Multisource and multitemporal data fusion in remote sensing a comprehensive review of the state of the art. Ieee Geoscience and Remote Sensing Magazine, 7(1), 6–39. https://doi.org/10.1109/Mgrs.2018.2890023
Ghulam, A., Ghulam, O., Maimaitijiang, M., Freeman, K., Porton, I., & Maimaitiyiming, M. (2015). Remote sensing based spatial statistics to document tropical rainforest transition pathways. Remote Sensing, 7(5), 6257–6279. https://doi.org/10.3390/rs70506257
Gopal, P. S. M., & Bhargavi, R. (2019). A novel approach for efficient crop yield prediction. Computers and Electronics and Agriculture. https://doi.org/10.1016/j.compag.2019.104968
Guo, A., Ye, H., Li, G., Zhang, B., Huang, W., Jiao, Q., Qian, B., & Luo, P. (2023). Evaluation of hybrid models for maize chlorophyll retrieval using medium- and high-spatial-resolution satellite images. Remote Sensing, 15(7), 1784. https://doi.org/10.3390/rs15071784
Haghighattalab, A., Crain, J., Mondal, S., Rutkoski, J., Singh, R. P., & Poland, J. (2017). Application of geographically weighted regression to improve grain yield prediction from unmanned aerial system imagery. Crop Science, 57(5), 2478–2489. https://doi.org/10.2135/cropsci2016.12.1016
Han, J. C., Zhang, Z., Cao, J., Luo, Y. C., Zhang, L. L., Li, Z. Y., & Zhang, J. (2020). Prediction of winter wheat yield based on multi-source data and machine learning in China. Remote Sensing. https://doi.org/10.3390/rs12020236
Hara, P., Piekutowska, M., & Niedbala, G. (2021). Selection of independent variables for crop yield prediction using artificial neural network models with remote sensing data. Land, 10(6), 609. https://doi.org/10.3390/land10060609
Harries, K. (2006). Extreme spatial variations in crime density in Baltimore County, MD. Geoforum, 37(3), 404–416. https://doi.org/10.1016/j.geoforum.2005.09.004
Hassan, M. A., Yang, M. J., Rasheed, A., Yang, G. J., Reynolds, M., Xia, X. C., Xiao, Y. G., & He, Z. H. (2019). A rapid monitoring of NDVI across the wheat growth cycle for grain yield prediction using a multi-spectral UAV platform. Plant Science, 282, 95–103. https://doi.org/10.1016/j.plantsci.2018.10.022
Hauglin, M., & Orka, H. O. (2016). Discriminating between native Norway Spruce and invasive Sitka Spruce-a comparison of multitemporal landsat 8 imagery, aerial images and airborne laser scanner data. Remote Sensing. https://doi.org/10.3390/rs8050363
He, K., X. Zhang, S. Ren, & J. Sun. (2016). “Deep Residual Learning for Image Recognition.” 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 27–30 June 2016.
Hoeser, T., & Kuenzer, C. (2020). Object detection and image segmentation with deep learning on earth observation data: A review-part I: evolution and recent trends. Remote Sensing. https://doi.org/10.3390/rs12101667
Hu, H. F., Scheben, A., & Edwards, D. (2018). Advances in integrating genomics and bioinformatics in the plant breeding pipeline. Agriculture-Basel. https://doi.org/10.3390/agriculture8060075
Huang, G., Z. Liu, L. V. D. Maaten, & K. Q. Weinberger. (2017). “Densely Connected Convolutional Networks.” 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 21–26 July 2017.
Huang, J., Gomez-Dans, J. L., Huang, H., Ma, H., Wu, Q., Lewis, P. E., Liang, S., Chen, Z., Xue, J., Wu, Y., Zhao, F., Wang, J., & Xie, X. (2019). Assimilation of remote sensing into crop growth models: Current status and perspectives. Agricultural and Forest Meteorology. https://doi.org/10.1016/j.agrformet.2019.06.008
Hussain, M., Bird, J. J., & Faria, D. R. (2019). A study on CNN transfer learning for image classification. In A. Lotfi, H. Bouchachia, A. Gegov, C. Langensiepen, & M. McGinnity (Eds.), Advances in computational intelligence systems (pp. 191–202). Springer.
Ide, H., & T. Kurita. (2017). “Improvement of learning for CNN with ReLU activation by sparse regularization.” 2017 International Joint Conference on Neural Networks (IJCNN), 14–19 May 2017.
Iqbal, F., Lucieer, A., & Barry, K. (2018). Simplified radiometric calibration for UAS-mounted multispectral sensor. European Journal of Remote Sensing, 51(1), 301–313. https://doi.org/10.1080/22797254.2018.1432293
Jain, M., Solomon, D., Capnerhurst, H., Arnold, A., Elliott, A., Kinzer, A. T., Knauss, C., Peters, M., Rolf, B., Weil, A., & Weinstein, C. (2020). How much can sustainable intensification increase yields across South Asia? A systematic review of the evidence. Environmental Research Letters. https://doi.org/10.1088/1748-9326/ab8b10
Jeong, J. H., Resop, J. P., Mueller, N. D., Fleisher, D. H., Yun, K., Butler, E. E., Timlin, D. J., Shim, K. M., Gerber, J. S., Reddy, V. R., & Kim, S. H. (2016). Random forests for global and regional crop yield predictions. PLoS ONE. https://doi.org/10.1371/journal.pone.0156571
Ji, S. P., Zhang, C., Xu, A. J., Shi, Y., & Duan, Y. L. (2018). 3D convolutional neural networks for crop classification with multi-temporal remote sensing images. Remote Sensing. https://doi.org/10.3390/rs10010075
Kasampalis, D. A., Alexandridis, T. K., Deva, C., Challinor, A., Moshou, D., & Zalidis, G. (2018). Contribution of remote sensing on crop models: A review. Journal of Imaging. https://doi.org/10.3390/jimaging4040052
Khaki, A., Pham, H., & Wang, L. (2021). Simultaneous corn and soybean yield prediction from remote sensing data using deep transfer learning. Scientific Reports. https://doi.org/10.1038/s41598-021-89779-z
Kingma, D. P., & J. Ba. (2015). “Adam: A Method for Stochastic Optimization.” 3rd International Conference for Learning Representations, San Diego.
Kipkulei, H. K., Bellingrath-Kimura, S. D., Lana, M., Ghazaryan, G., Baatz, R., Boitt, M., Chisanga, C. B., Rotich, B., & Sieber, S. (2022). Assessment of maize yield response to agricultural management strategies using the DSSAT–CERES-maize model in Trans Nzoia County in Kenya. International Journal of Plant Production, 16, 557–577. https://doi.org/10.1007/s42106-022-00220-5
Knyazikhin, Y., Schull, M. A., Stenberg, P., Mottus, M., Rautiainen, M., Yang, Y., Marshak, A., Carmona, P. L., Kaufmann, R. K., Lewis, P., Disney, M. I., Vanderbilt, V., Davis, A. B., Baret, F., Jacquemoud, S., Lyapustin, A., & Myneni, R. B. (2013). Hyperspectral remote sensing of foliar nitrogen content. Proceedings of the National Academy of Sciences of the United States of America, 110(3), E185–E192. https://doi.org/10.1073/pnas.1210196109
Kogan, F., Kussul, N., Adamenko, T., Skakun, S., Kravchenko, O., Kryvobok, O., Shelestov, A., Kolotii, A., Kussul, O., & Lavrenyuk, A. (2013). Winter wheat yield forecasting in Ukraine based on Earth observation, meteorological data and biophysical models. International Journal of Applied Earth Observation and Geoinformation, 23, 192–203. https://doi.org/10.1016/j.jag.2013.01.002
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2017). ImageNet classification with deep convolutional neural networks. Communications of the Acm, 60(6), 84–90. https://doi.org/10.1145/3065386
Kumar, L., & Mutanga, O. (2017). Remote sensing of above-ground biomass. Remote Sensing. https://doi.org/10.3390/rs9090935
Li, B., Xu, X. M., Zhang, L., Han, J. W., Bian, C. S., Li, G. C., Liu, J. G., & Jin, L. P. (2020). Above-ground biomass estimation and yield prediction in potato by using UAV-based RGB and hyperspectral imaging. Isprs Journal of Photogrammetry and Remote Sensing, 162, 161–172. https://doi.org/10.1016/j.isprsjprs.2020.02.013
Li, Z., Chen, Z., Cheng, Q., Fei, S., & Zhou, X. (2023). Deep learning models outperform generalized machine learning models in predicting winter wheat yield based on multispectral data from drones. Drones. https://doi.org/10.3390/drones7080505
Lobell, D. B., Cassman, K. G., & Field, C. B. (2009). Crop yield gaps: their importance, magnitudes, and causes. Annual Review of Environment and Resources, 34, 179–204. https://doi.org/10.1146/annurev.environ.041008.093740
Lu, D. S., Chen, Q., Wang, G. X., Liu, L. J., Li, G. Y., & Moran, E. (2016). A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. International Journal of Digital Earth, 9(1), 63–105. https://doi.org/10.1080/17538947.2014.990526
Maimaitijiang, M., Ghulam, A., Sandoval, J. S. O., & Maimaitiyiming, M. (2015). Drivers of land cover and land use changes in St. Louis metropolitan area over the past 40 years characterized by remote sensing and census population data. International Journal of Applied Earth Observation and Geoinformation, 35, 161–174. https://doi.org/10.1016/j.jag.2014.08.020
Maimaitijiang, M., Ghulam, A., Sidike, P., Hartling, S., Maimaitiyiming, M., Peterson, K., Shavers, E., Fishman, J., Peterson, J., Kadam, S., Burken, J., & Fritschi, F. (2017). Unmanned Aerial System (UAS)-based phenotyping of soybean using multi-sensor data fusion and extreme learning machine. ISPRS Journal of Photogrammetry and Remote Sensing, 134, 43–58. https://doi.org/10.1016/j.isprsjprs.2017.10.011
Maimaitijiang, M., Sagan, V., Sidike, P., Hartling, S., Esposito, F., & Fritschi, F. B. (2020). Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sensing of Environment. https://doi.org/10.1016/j.rse.2019.111599
Marsh, J. I., Hu, H. F., Gill, M., Batley, J., & Edwards, D. (2021). Crop breeding for a changing climate: Integrating phenomics and genomics with bioinformatics. Theoretical and Applied Genetics, 134(6), 1677–1690. https://doi.org/10.1007/s00122-021-03820-3
Mcmichael, A. J. (1994). Global environmental-change and human health - new challenges to scientist and policy-maker. Journal of Public Health Policy, 15(4), 407–419. https://doi.org/10.2307/3343023
Morales, G., Sheppard, J. W., Hegedus, P. B., & Maxwell, B. (2023). Improved yield prediction of winter wheat using a novel two-dimensional deep regression neural network trained via remote sensing. Sensors, 23(1), 489. https://doi.org/10.3390/s23010489
Nair, V., & G. E. Hinton. (2010). Rectified Linear Units Improve Restricted Boltzmann Machines. Proceedings of the 27th International Conference on International Conference on Machine Learning: 807–814. https://doi.org/10.5555/3104322.3104425.
Nanni, L., Ghidoni, S., & Brahnam, S. (2017). Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recognition, 71, 158–172. https://doi.org/10.1016/j.patcog.2017.05.025
Nevavuori, P., Narra, N., Linna, P., & Lipping, T. (2020). Crop yield prediction using multitemporal uav data and spatio-temporal deep learning models. Remote Sensing. https://doi.org/10.3390/rs12234000
Nevavuori, P., Narra, N., & Lipping, T. (2019). Crop yield prediction with deep convolutional neural networks. Computers and Electronics in Agriculture. https://doi.org/10.1016/j.compag.2019.104859
Papageorgiou, E. I., Aggelopoulou, K. D., Gemtos, T. A., & Nanos, G. D. (2013). Yield prediction in apples using fuzzy cognitive map learning approach. Computers and Electronics in Agriculture, 91, 19–29. https://doi.org/10.1016/j.compag.2012.11.008
Papageorgiou, E. I., Markinos, A. T., & Gemtos, T. A. (2011). Fuzzy cognitive map based approach for predicting yield in cotton crop production as a basis for decision support system in precision agriculture application. Applied Soft Computing, 11(4), 3643–3657. https://doi.org/10.1016/j.asoc.2011.01.036
Peng, B., Guan, K. Y., Zhou, W., Jiang, C. Y., Frankenberg, C., Sun, Y., He, L. Y., & Kohler, P. (2020). Assessing the benefit of satellite-based solar-induced chlorophyll fluorescence in crop yield prediction. International Journal of Applied Earth Observation and Geoinformation. https://doi.org/10.1016/j.jag.2020.102126
Peralta, N. R., Assefa, Y., Du, J., Barden, C. J., & Ciampitti, I. A. (2016). Mid-season high-resolution satellite imagery for forecasting site-specific corn yield. Remote Sensing. https://doi.org/10.3390/rs8100848
Rischbeck, P., Elsayed, S., Mistele, B., Barmeier, G., Heil, K., & Schmidhalter, U. (2016). Data fusion of spectral, thermal and canopy height parameters for improved yield prediction of drought stressed spring barley. European Journal of Agronomy, 78, 44–59. https://doi.org/10.1016/j.eja.2016.04.013
Sagan, V., Maimaitijiang, M., Bhadra, S., Maimaitiyiming, M., Brown, D. R., Sidike, P., & Fritschi, F. B. (2021). Field-scale crop yield prediction using multi-temporal WorldView-3 and PlanetScope satellite data and deep learning. Isprs Journal of Photogrammetry and Remote Sensing, 174, 265–281. https://doi.org/10.1016/j.isprsjprs.2021.02.008
Schwalbert, R. A., Amado, T., Corassa, G., Pott, L. P., Prasad, P. V. V., & Ciampitti, I. A. (2020). Satellite-based soybean yield forecast: Integrating machine learning and weather data for improving crop yield prediction in southern Brazil. Agricultural and Forest Meteorology. https://doi.org/10.1016/j.agrformet.2019.107886
Shakoor, N., Lee, S., & Mockler, T. C. (2017). High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Current Opinion in Plant Biology, 38, 184–192. https://doi.org/10.1016/j.pbi.2017.05.006
Shen, Y., Mercatoris, B., Cao, Z., Kwan, P., Guo, L., Yaoi, H., & Cheng, Q. (2022). Improving wheat yield prediction accuracy using LSTM-RF framework based on UAV thermal infrared and multispectral imagery. Agriculture. https://doi.org/10.3390/agriculture12060892
Simonyan, K., & A. Zisserman. (2015). “Very Deep Convolutional Networks for Large-Scale Image Recognition.” International Conference on Learning Representations, San Diego.
Singh, S. P., Wang, L. P., Gupta, S., Goli, H., Padmanabhan, P., & Gulyas, B. (2020). 3D deep learning on medical images: A review. Sensors. https://doi.org/10.3390/s20185097
Song, P., Wang, J. L., Guo, X. Y., Yang, W. N., & Zhao, C. J. (2021). High-throughput phenotyping: Breaking through the bottleneck in future crop breeding. Crop Journal, 9(3), 633–645. https://doi.org/10.1016/j.cj.2021.03.015
Su, S., C. Zhang, K. Han, and Y. Tian. (2018). Greedy Hash: Towards Fast Optimization for Accurate Hash Coding in CNN. Advances in Neural Information Processing Systems 31. https://proceedings.neurips.cc/paper/2018/file/13f3cf8c531952d72e5847c4183e6910-Paper.pdf.
Sun, J., Di, L. P., Sun, Z. H., Shen, Y. L., & Lai, Z. L. (2019). County-level soybean yield prediction using deep CNN-LSTM model. Sensors. https://doi.org/10.3390/s19204363
Sun, Z. H., Di, L. P., Fang, H., & Burgess, A. (2020). Deep learning classification for crop types in North Dakota. Ieee Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 13, 2200–2213. https://doi.org/10.1109/Jstars.2020.2990104
Suzuki, K. (2017). Overview of deep learning in medical imaging. Radiological Physics and Technology, 10(3), 257–273. https://doi.org/10.1007/s12194-017-0406-5
Tan, C., F. Sun, T. Kong, W. Zhang, C. Yang, and C. Liu. (2018). “A Survey on Deep Transfer Learning.” Artificial Neural Networks and Machine Learning – ICANN 2018, Cham, 2018//.
Tanabe, R., Matsui, T., & Tanaka, T. S. T. (2023). Winter wheat yield prediction using convolutional neural networks and UAV-based multispectral imagery. Field Crops Research. https://doi.org/10.1016/j.fcr.2022.108786
van Dijk, M., Morley, T., Rau, M. L., & Saghai, Y. (2021). A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nature Food, 2(7), 494. https://doi.org/10.1038/s43016-021-00322-9
van Klompenburg, T., Kassahun, A., & Catal, C. (2020). Crop yield prediction using machine learning: A systematic literature review. Computers and Electronics in Agriculture. https://doi.org/10.1016/j.compag.2020.105709
Wallace, L. (2013). “Assessing the stability of canopy maps produced from UAV-LiDAR data.” 2013 IEEE International Geoscience and Remote Sensing Symposium - IGARSS, 21–26 July 2013.
Wan, L., Cen, H. Y., Zhu, J. P., Zhang, J. F., Zhu, Y. M., Sun, D. W., Du, X. Y., Zhai, L., Weng, H. Y., Li, Y. J., Li, X. R., Bao, Y. D., Shou, J. Y., & He, Y. (2020). Grain yield prediction of rice using multi-temporal UAV-based RGB and multispectral images and model transfer - a case study of small farmlands in the South of China. Agricultural and Forest Meteorology. https://doi.org/10.1016/j.agrformet.2020.108096
Wang, L. A., Zhou, X. D., Zhu, X. K., & Guo, W. S. (2017). Estimation of leaf nitrogen concentration in wheat using the MK-SVR algorithm and satellite remote sensing data. Computers and Electronics in Agriculture, 140, 327–337. https://doi.org/10.1016/j.compag.2017.05.023
Wheeler, T., & von Braun, J. (2013). Climate change impacts on global food security. Science, 341(6145), 508–513. https://doi.org/10.1126/science.1239402
Xie, S., R. Girshick, P. Dollár, Z. Tu, and K. He. (2017). “Aggregated Residual Transformations for Deep Neural Networks.” 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 21–26 July 2017.
Xie, H., Hicks, N., Keller, G. R., Huang, H., & Kreinovich, V. (2003). An IDL/ENVI implementation of the FFT-based algorithm for automatic image registration. Computers & Geosciences, 29(8), 1045–1055. https://doi.org/10.1016/S0098-3004(03)00104-3
Xie, Q. Y., Dash, A. D., Huete, A. R. O., Jiang, A. H., Yin, G. F., Ding, Y. L., Peng, D. L., Hall, R. O. E., Brown, L. K., Shi, Y., Ye, H. C., Dong, Y. Y., & Huang, W. J. (2019). Retrieval of crop biophysical parameters from sentinel-2 remote sensing imagery. International Journal of Applied Earth Observation and Geoinformation, 80, 187–195. https://doi.org/10.1016/j.jag.2019.04.019
Yang, M. M., Ma, T. H., Tian, Q., Tian, Y., Al-Dhelaan, A., & Al-Dhelaan, M. (2022a). Aggregated squeeze-and-excitation transformations for densely connected convolutional networks. Visual Computer, 38(8), 2661–2674. https://doi.org/10.1007/s00371-021-02144-z
Yang, Q., Shi, L. S., Han, J. Y., Zha, Y. Y., & Zhu, P. H. (2019). Deep convolutional neural networks for rice grain yield estimation at the ripening stage using UAV-based remotely sensed images. Field Crops Research, 235, 142–153. https://doi.org/10.1016/j.fcr.2019.02.022
Yang, Y., Hu, Y. Y., Zhang, X. Y., & Wang, S. (2022b). Two-stage selective ensemble of CNN via deep tree training for medical image classification. Ieee Transactions on Cybernetics, 52(9), 9194–9207. https://doi.org/10.1109/Tcyb.2021.3061147
Zeiler, M. D., and R. Fergus. (2014). “Visualizing and Understanding Convolutional Networks.” Computer Vision – ECCV 2014, Cham, 2014.
Zhou, X., Zheng, H. B., Xu, X. Q., He, J. Y., Ge, X. K., Yao, X., Cheng, T., Zhu, Y., Cao, W. X., & Tian, Y. C. (2017). Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multispectral and digital imagery. ISPRS Journal of Photogrammetry and Remote Sensing, 130, 246–255. https://doi.org/10.1016/j.isprsjprs.2017.05.003
Zhuang, F., Qi, Z., Duan, K., Xi, D., Zhu, Y., Zhu, H., Xiong, H., & He, Q. (2021). A comprehensive survey on transfer learning. Proceedings of the IEEE, 109(1), 43–76. https://doi.org/10.1109/JPROC.2020.3004555
Zhuo, W., Fang, S. B., Gao, X. R., Wang, L., Wu, D., Fu, S. L., Wu, Q. L., & Huang, J. X. (2022). Crop yield prediction using MODIS LAI, TIGGE weather forecasts and WOFOST model: A case study for winter wheat in Hebei, China during 2009–2013. International Journal of Applied Earth Observation and Geoinformation. https://doi.org/10.1016/j.jag.2021.102668
Zhuo, W., Huang, J. X., Li, L., Zhang, X. D., Ma, H. Y., Gao, X. R., Huang, H., Xu, B. D., & Xiao, X. M. (2019). Assimilating soil moisture retrieved from sentinel-1 and sentinel-2 data into WOFOST model to improve winter wheat yield estimation. Remote Sensing. https://doi.org/10.3390/rs11131618
Acknowledgements
This work was supported in part by GDM Seeds, United Soybean Board Project #: 2331-201-0103, 2431-201-0101 and in part by USGS AmericaView Grant (G18AP00077). Additional financial and computing support was provided by the Taylor Geospatial Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest regarding this research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bhadra, S., Sagan, V., Skobalski, J. et al. End-to-end 3D CNN for plot-scale soybean yield prediction using multitemporal UAV-based RGB images. Precision Agric 25, 834–864 (2024). https://doi.org/10.1007/s11119-023-10096-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-023-10096-8