当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Uniform Molecular Low-Density Parity Check Decoder.
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-12-18 , DOI: 10.1021/acssynbio.8b00304 Chuan Zhang , Lulu Ge , Xingchi Zhang , Wei Wei , Jing Zhao , Zaichen Zhang , Zhongfeng Wang , Xiaohu You
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-12-18 , DOI: 10.1021/acssynbio.8b00304 Chuan Zhang , Lulu Ge , Xingchi Zhang , Wei Wei , Jing Zhao , Zaichen Zhang , Zhongfeng Wang , Xiaohu You
|
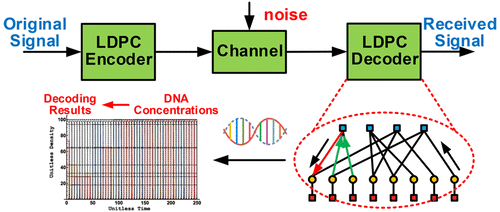
Error correction codes, such as low-density parity check (LDPC) codes, are required to be larger scale to meet the increasing demands for reliable and massive data transmission. However, the construction of such a large-scale decoder will result in high complexity and hinder its silicon implementation. Thanks to the advantages of natural computing in high parallelism and low power, we propose a method to synthesize a uniform molecular LDPC decoder by implementing the belief-propagation algorithm with chemical reaction networks (CRNs). This method enables us to flexibly design the LDPC decoder with arbitrary code length, code rate, and node degrees. Compared with existing methods, our proposal reduces the number of reactions to update the variable nodes by 42.86% and the check nodes by 47.37%. Numerical results are presented to show the feasibility and validity of our proposal.
中文翻译:

统一的分子低密度奇偶校验解码器。
诸如低密度奇偶校验(LDPC)码之类的纠错码需要更大的规模,以满足对可靠,大量数据传输不断增长的需求。然而,这种大规模解码器的构造将导致高复杂度并阻碍其硅实现。由于自然计算具有高并行性和低功耗的优点,我们提出了一种通过使用化学反应网络(CRN)实现置信度传播算法来合成均匀分子LDPC解码器的方法。这种方法使我们能够灵活地设计具有任意代码长度,代码率和节点度的LDPC解码器。与现有方法相比,我们的建议将更新变量节点的反应数量减少了42.86%,将校验节点的反应数量减少了47.37%。
更新日期:2018-12-04
中文翻译:

统一的分子低密度奇偶校验解码器。
诸如低密度奇偶校验(LDPC)码之类的纠错码需要更大的规模,以满足对可靠,大量数据传输不断增长的需求。然而,这种大规模解码器的构造将导致高复杂度并阻碍其硅实现。由于自然计算具有高并行性和低功耗的优点,我们提出了一种通过使用化学反应网络(CRN)实现置信度传播算法来合成均匀分子LDPC解码器的方法。这种方法使我们能够灵活地设计具有任意代码长度,代码率和节点度的LDPC解码器。与现有方法相比,我们的建议将更新变量节点的反应数量减少了42.86%,将校验节点的反应数量减少了47.37%。



























 京公网安备 11010802027423号
京公网安备 11010802027423号