Nature Methods ( IF 48.0 ) Pub Date : 2018-11-26 , DOI: 10.1038/s41592-018-0214-9 Rachelly Normand 1 , Wenfei Du 2 , Mayan Briller 1 , Renaud Gaujoux 1 , Elina Starosvetsky 1 , Amit Ziv-Kenet 1 , Gali Shalev-Malul 1 , Robert J Tibshirani 2, 3 , Shai S Shen-Orr 1
|
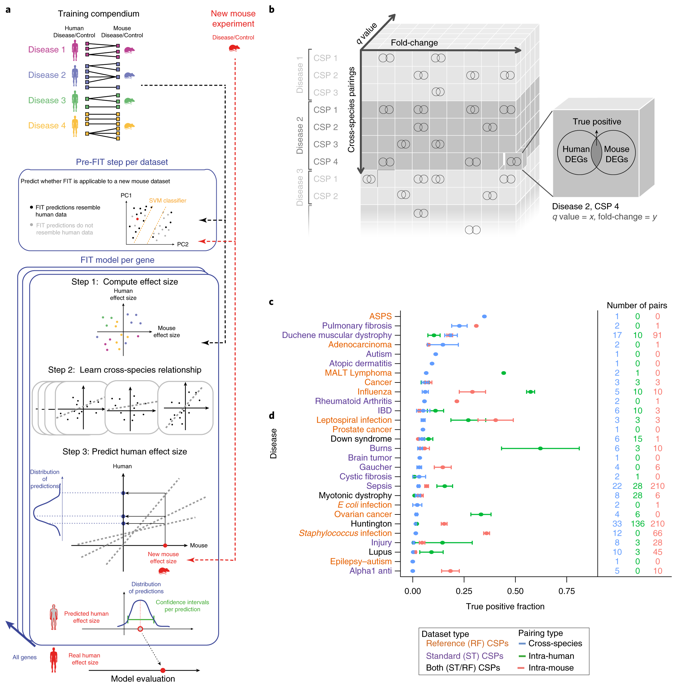
Cross-species differences form barriers to translational research that ultimately hinder the success of clinical trials, yet knowledge of species differences has yet to be systematically incorporated in the interpretation of animal models. Here we present Found In Translation (FIT; http://www.mouse2man.org), a statistical methodology that leverages public gene expression data to extrapolate the results of a new mouse experiment to expression changes in the equivalent human condition. We applied FIT to data from mouse models of 28 different human diseases and identified experimental conditions in which FIT predictions outperformed direct cross-species extrapolation from mouse results, increasing the overlap of differentially expressed genes by 20–50%. FIT predicted novel disease-associated genes, an example of which we validated experimentally. FIT highlights signals that may otherwise be missed and reduces false leads, with no experimental cost.
中文翻译:

Found In Translation:一种用于人机推理的机器学习模型
跨物种差异构成了转化研究的障碍,最终阻碍了临床试验的成功,但物种差异的知识尚未系统地纳入动物模型的解释中。在这里,我们介绍了 Found In Translation (FIT;http://www.mouse2man.org),这是一种统计方法,它利用公共基因表达数据将新小鼠实验的结果外推到同等人类条件下的表达变化。我们将 FIT 应用于来自 28 种不同人类疾病的小鼠模型的数据,并确定了 FIT 预测优于从小鼠结果直接跨物种外推的实验条件,将差异表达基因的重叠增加了 20-50%。FIT 预测了新的疾病相关基因,这是我们通过实验验证的一个例子。


























 京公网安备 11010802027423号
京公网安备 11010802027423号