当前位置:
X-MOL 学术
›
Nat. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Quantifying and comparing bacterial growth dynamics in multiple metagenomic samples.
Nature Methods ( IF 48.0 ) Pub Date : 2018-11-12 , DOI: 10.1038/s41592-018-0182-0 Yuan Gao 1 , Hongzhe Li 1
Nature Methods ( IF 48.0 ) Pub Date : 2018-11-12 , DOI: 10.1038/s41592-018-0182-0 Yuan Gao 1 , Hongzhe Li 1
Affiliation
|
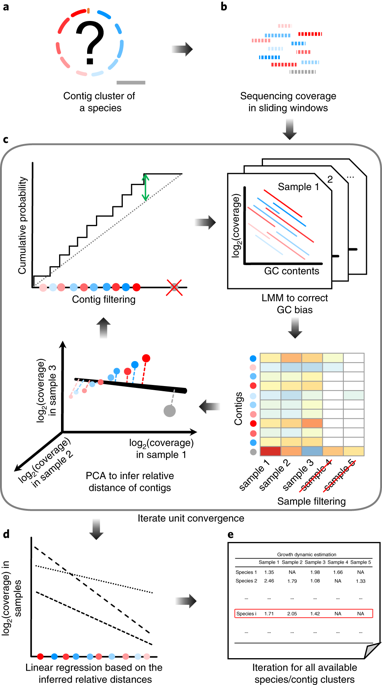
The accurate quantification of microbial growth dynamics for species without complete genome sequences is biologically important, but computationally challenging in metagenomics. Here we present dynamic estimator of microbial communities (DEMIC; https://sourceforge.net/projects/demic/ ), a multi-sample algorithm based on contigs and coverage values, to infer the relative distances of contigs from the replication origin and to accurately compare bacterial growth rates between samples. We demonstrate robust performances of DEMIC for various sample sizes and assembly qualities using multiple synthetic and real datasets.
中文翻译:

量化和比较多个宏基因组样本中细菌的生长动力学。
没有完整的基因组序列的物种的微生物生长动力学的准确定量在生物学上很重要,但是在宏基因组学上在计算上具有挑战性。在这里,我们介绍了微生物群落的动态估算器(DEMIC; https://sourceforge.net/projects/demic/),这是一种基于重叠群和覆盖率值的多样本算法,用以推断重叠群与复制起点之间的相对距离,以及准确比较样品之间的细菌生长速率。我们使用多个合成和真实数据集展示了DEMIC在各种样本大小和装配质量下的强大性能。
更新日期:2018-12-10
中文翻译:

量化和比较多个宏基因组样本中细菌的生长动力学。
没有完整的基因组序列的物种的微生物生长动力学的准确定量在生物学上很重要,但是在宏基因组学上在计算上具有挑战性。在这里,我们介绍了微生物群落的动态估算器(DEMIC; https://sourceforge.net/projects/demic/),这是一种基于重叠群和覆盖率值的多样本算法,用以推断重叠群与复制起点之间的相对距离,以及准确比较样品之间的细菌生长速率。我们使用多个合成和真实数据集展示了DEMIC在各种样本大小和装配质量下的强大性能。


























 京公网安备 11010802027423号
京公网安备 11010802027423号