当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Comprehensive Profiling of Four Base Overhang Ligation Fidelity by T4 DNA Ligase and Application to DNA Assembly
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-10-18 00:00:00 , DOI: 10.1021/acssynbio.8b00333 Vladimir Potapov 1 , Jennifer L. Ong 1 , Rebecca B. Kucera 2 , Bradley W. Langhorst 2 , Katharina Bilotti 1 , John M. Pryor 1 , Eric J. Cantor 2 , Barry Canton 3 , Thomas F. Knight 3 , Thomas C. Evans 1 , Gregory J. S. Lohman 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-10-18 00:00:00 , DOI: 10.1021/acssynbio.8b00333 Vladimir Potapov 1 , Jennifer L. Ong 1 , Rebecca B. Kucera 2 , Bradley W. Langhorst 2 , Katharina Bilotti 1 , John M. Pryor 1 , Eric J. Cantor 2 , Barry Canton 3 , Thomas F. Knight 3 , Thomas C. Evans 1 , Gregory J. S. Lohman 1
Affiliation
|
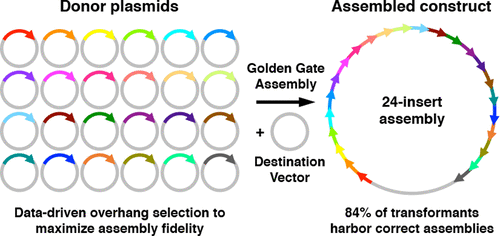
Synthetic biology relies on the manufacture of large and complex DNA constructs from libraries of genetic parts. Golden Gate and other Type IIS restriction enzyme-dependent DNA assembly methods enable rapid construction of genes and operons through one-pot, multifragment assembly, with the ordering of parts determined by the ligation of Watson–Crick base-paired overhangs. However, ligation of mismatched overhangs leads to erroneous assembly, and low-efficiency Watson Crick pairings can lead to truncated assemblies. Using sets of empirically vetted, high-accuracy junction pairs avoids this issue but limits the number of parts that can be joined in a single reaction. Here, we report the use of comprehensive end-joining ligation fidelity and bias data to predict high accuracy junction sets for Golden Gate assembly. The ligation profile accurately predicted junction fidelity in ten-fragment Golden Gate assembly reactions and enabled accurate and efficient assembly of a lac cassette from up to 24-fragments in a single reaction.
中文翻译:

T4 DNA连接酶对四碱基突出端连接保真度的综合分析及其在DNA组装中的应用
合成生物学依赖于从遗传部分的文库中制备大型和复杂的DNA构建体。金门(Golden Gate)和其他类型的IIS限制性内切酶依赖性DNA组装方法可通过一锅多片段组装快速构建基因和操纵子,其排列顺序由Watson-Crick碱基对突出端的连接决定。但是,不匹配突出端的连接会导致组装错误,而低效率的Watson Crick配对可能会导致组装被截断。使用一组经过经验验证的高精度结对可以避免此问题,但会限制单个反应中可以连接的部分的数量。在这里,我们报告了使用全面的末端连接连接保真度和偏倚数据来预测金门大会装配的高精度连接集。一个反应中最多可容纳24个片段的lac盒。
更新日期:2018-10-18
中文翻译:

T4 DNA连接酶对四碱基突出端连接保真度的综合分析及其在DNA组装中的应用
合成生物学依赖于从遗传部分的文库中制备大型和复杂的DNA构建体。金门(Golden Gate)和其他类型的IIS限制性内切酶依赖性DNA组装方法可通过一锅多片段组装快速构建基因和操纵子,其排列顺序由Watson-Crick碱基对突出端的连接决定。但是,不匹配突出端的连接会导致组装错误,而低效率的Watson Crick配对可能会导致组装被截断。使用一组经过经验验证的高精度结对可以避免此问题,但会限制单个反应中可以连接的部分的数量。在这里,我们报告了使用全面的末端连接连接保真度和偏倚数据来预测金门大会装配的高精度连接集。一个反应中最多可容纳24个片段的lac盒。


























 京公网安备 11010802027423号
京公网安备 11010802027423号