Chem ( IF 23.5 ) Pub Date : 2018-10-18 , DOI: 10.1016/j.chempr.2018.09.015 Pablo D. Dans , Diego Gallego , Alexandra Balaceanu , Leonardo Darré , Hansel Gómez , Modesto Orozco
|
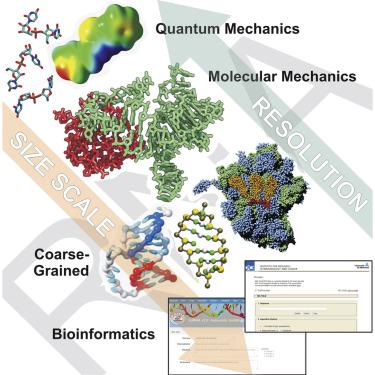
Although chemically close to DNA, RNA can adopt a wide range of structures from regular helices to complex globular conformations, showing a complexity similar to that of proteins. The determination of the structure of RNA molecules, crucial for functional understanding, is severely handicapped by their size and flexibility, which makes the systematic use of experimental approaches difficult. Simulation techniques are also suffering from very severe problems related to the accuracy of the methods and their ability to sample a large and complex conformational landscape. Here, we systematically review recent approaches created to reduce the shortcoming of the current generation of simulation methods—from highly accurate models able to deal with small systems to coarse-grained approaches that are less accurate but applicable to dealing with large models.
中文翻译:

RNA结构的建模,仿真和生物信息学
尽管化学上接近DNA,但RNA可以采用多种结构,从规则的螺旋到复杂的球状构象,显示出与蛋白质相似的复杂性。RNA分子结构的确定对于功能性理解至关重要,但由于其大小和灵活性而受到严重阻碍,这使得系统地使用实验方法变得困难。模拟技术还面临着与方法的准确性及其对大而复杂的构象景观进行采样的能力有关的非常严重的问题。这里,



























 京公网安备 11010802027423号
京公网安备 11010802027423号