当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Selective Elimination of the Key Subunit Interfaces Facilitates Conversion of Native 24-mer Protein Nanocage into 8-mer Nanorings
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2018-10-17 , DOI: 10.1021/jacs.8b09760 Wenming Wang 1 , Lele Wang 1 , Hai Chen 2 , Jiachen Zang 2 , Xuan Zhao 1 , Guanghua Zhao 2 , Hongfei Wang 1
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2018-10-17 , DOI: 10.1021/jacs.8b09760 Wenming Wang 1 , Lele Wang 1 , Hai Chen 2 , Jiachen Zang 2 , Xuan Zhao 1 , Guanghua Zhao 2 , Hongfei Wang 1
Affiliation
|
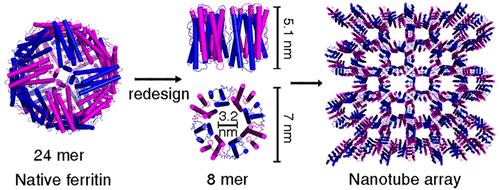
Living systems utilize proteins as building blocks to construct a large variety of self-assembled nanoscale architectures. Yet, creating protein-based assemblies with specific geometries in the laboratory remains challenging. Here, we present a new approach that completely eliminates one natural intersubunit interface of multisubunit protein architecture with high symmetry, resulting in reassembly of the protein architecture into one with lower symmetry. We have applied this approach to the conversion of the 24-mer cage-like ferritin into non-native 8-mer protein nanorings in solution. In the crystal structure, such newly formed nanorings connect with each other through hydrogen bonding in a repeating head-to-tail pattern to form nanotubes, and adjacent nanotubes are staggered relative to one another to create three-dimensional porous protein assemblies. The above strategy allows the study of conversion between protein architectures with different geometries by adjusting the interactions at the intersubunit interfaces, and the fabrication of novel bio-nanomaterials with different geometries.
中文翻译:

选择性消除关键亚基界面促进天然 24 聚体蛋白质纳米笼转化为 8 聚体纳米环
生命系统利用蛋白质作为构建块来构建各种各样的自组装纳米级结构。然而,在实验室中创建具有特定几何形状的基于蛋白质的组件仍然具有挑战性。在这里,我们提出了一种新方法,它完全消除了具有高度对称性的多亚基蛋白质结构的一个天然亚基间界面,从而将蛋白质结构重新组装成具有较低对称性的结构。我们已将这种方法应用于将 24 聚体笼状铁蛋白在溶液中转化为非天然 8 聚体蛋白纳米环。在晶体结构中,这种新形成的纳米环通过氢键以重复的头对尾模式相互连接,形成纳米管,和相邻的纳米管相对于彼此交错以创建三维多孔蛋白质组件。上述策略允许通过调整亚基间界面的相互作用来研究具有不同几何形状的蛋白质结构之间的转换,以及制造具有不同几何形状的新型生物纳米材料。
更新日期:2018-10-17
中文翻译:

选择性消除关键亚基界面促进天然 24 聚体蛋白质纳米笼转化为 8 聚体纳米环
生命系统利用蛋白质作为构建块来构建各种各样的自组装纳米级结构。然而,在实验室中创建具有特定几何形状的基于蛋白质的组件仍然具有挑战性。在这里,我们提出了一种新方法,它完全消除了具有高度对称性的多亚基蛋白质结构的一个天然亚基间界面,从而将蛋白质结构重新组装成具有较低对称性的结构。我们已将这种方法应用于将 24 聚体笼状铁蛋白在溶液中转化为非天然 8 聚体蛋白纳米环。在晶体结构中,这种新形成的纳米环通过氢键以重复的头对尾模式相互连接,形成纳米管,和相邻的纳米管相对于彼此交错以创建三维多孔蛋白质组件。上述策略允许通过调整亚基间界面的相互作用来研究具有不同几何形状的蛋白质结构之间的转换,以及制造具有不同几何形状的新型生物纳米材料。


























 京公网安备 11010802027423号
京公网安备 11010802027423号