当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Efficient Sampling and Characterization of Free Energy Landscapes of Ion–Peptide Systems
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-09-21 00:00:00 , DOI: 10.1021/acs.jctc.8b00560 Tobias Lemke 1 , Christine Peter 1 , Oleksandra Kukharenko 1
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-09-21 00:00:00 , DOI: 10.1021/acs.jctc.8b00560 Tobias Lemke 1 , Christine Peter 1 , Oleksandra Kukharenko 1
Affiliation
|
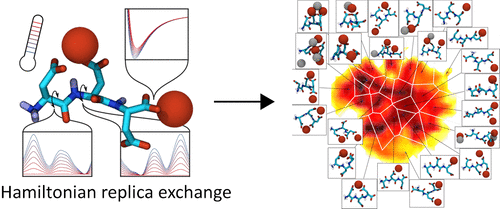
Proteins that influence nucleation, growth, or polymorph selection during biomineralization processes are often rich in glutamic- or aspartic acid. Here, the interactions between carboxylate side chains and ions lead to an interplay of peptide conformations and ion structuring in solution. Molecular dynamics simulations are an ideal tool to mechanistically investigate these processes. Unfortunately, the formation of strong ion-peptide contacts and ion bridges drastically impedes structural reorganization of ionic bonds and conformational transitions of the polymers. Thus, to obtain a complete thermodynamical picture of such systems, enhanced sampling techniques become necessary as well as the methods to characterize the conformational states of these partially disordered polymer-ion systems. Here, we propose a new set of Hamiltonian replica exchange (HRE) parameters for efficient simulations of peptide–ion systems, with an aspartic acid trimer in the presence of Ca2+ and Cl– ions as a test system. We introduce dimensionality reduction and clustering strategies to characterize the states of such a multicomponent system and to analyze the outcome of the proposed HRE with different reweighting methods.
中文翻译:

离子-肽系统自由能态的有效采样和表征
在生物矿化过程中影响成核,生长或多晶型选择的蛋白质通常富含谷氨酸或天冬氨酸。此处,羧酸根侧链与离子之间的相互作用导致溶液中肽构象与离子结构的相互作用。分子动力学模拟是机械研究这些过程的理想工具。不幸的是,牢固的离子-肽接触和离子桥的形成极大地阻碍了离子键的结构重组和聚合物的构象转变。因此,为了获得此类系统的完整热力学图,必须使用增强的采样技术以及表征这些部分无序的聚合物离子系统的构象状态的方法。这里,2+和Cl -离子作为测试系统。我们引入降维和聚类策略来表征这种多组件系统的状态,并使用不同的加权方法来分析所提出的HRE的结果。
更新日期:2018-09-21
中文翻译:

离子-肽系统自由能态的有效采样和表征
在生物矿化过程中影响成核,生长或多晶型选择的蛋白质通常富含谷氨酸或天冬氨酸。此处,羧酸根侧链与离子之间的相互作用导致溶液中肽构象与离子结构的相互作用。分子动力学模拟是机械研究这些过程的理想工具。不幸的是,牢固的离子-肽接触和离子桥的形成极大地阻碍了离子键的结构重组和聚合物的构象转变。因此,为了获得此类系统的完整热力学图,必须使用增强的采样技术以及表征这些部分无序的聚合物离子系统的构象状态的方法。这里,2+和Cl -离子作为测试系统。我们引入降维和聚类策略来表征这种多组件系统的状态,并使用不同的加权方法来分析所提出的HRE的结果。


























 京公网安备 11010802027423号
京公网安备 11010802027423号