当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Accelerated Construction of Kinetic Network Model of Biomolecules Using Steered Molecular Dynamics
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-09-13 00:00:00 , DOI: 10.1021/acs.jctc.8b00398 Susmita Ghosh 1 , Abhijit Chatterjee 2 , Swati Bhattacharya 2
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-09-13 00:00:00 , DOI: 10.1021/acs.jctc.8b00398 Susmita Ghosh 1 , Abhijit Chatterjee 2 , Swati Bhattacharya 2
Affiliation
|
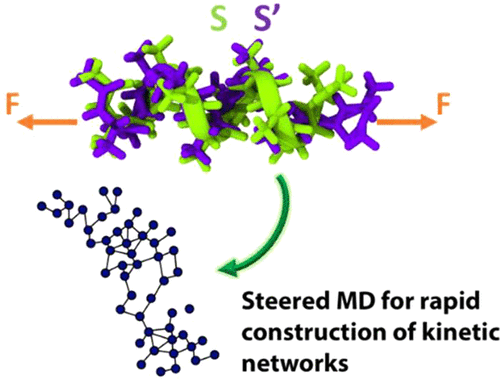
A new class of rare event acceleration techniques based on steered molecular dynamics (SMD) simulations is introduced. A stretching force applied on a biomolecule causes it to access large end-to-end distances. Under these conditions the biomolecule undergoes rapid conformational changes that are rare at zero-force conditions. A theory describing kinetics of a biomolecule at various stretching forces is presented. Using the theory, a master-Markov state model (master-MSM) is constructed from rates frequently accessed over a small range of force conditions. The master-MSM is shown to be applicable over a wide range of force conditions. We demonstrate application of the theory to three different biomolecular systems, namely, deca-alanine, TBA (thrombin binding aptamer), and a RNA hairpin. The master-MSM is used to estimate the kinetics at zero-force conditions, i.e., on the unbiased free-energy landscape, resulting inasmuch as 2–6 orders-of-magnitude speed-up over standard molecular dynamics.
中文翻译:

操纵分子动力学加速构建生物分子动力学网络模型
介绍了基于转向分子动力学(SMD)模拟的一类新的罕见事件加速技术。施加在生物分子上的拉伸力使其进入较大的端到端距离。在这些条件下,生物分子会经历快速构象变化,这在零力条件下是罕见的。提出了描述生物分子在各种拉伸力下的动力学的理论。使用该理论,可以从在很小的力条件范围内经常访问的速率构造主-马尔可夫状态模型(master-MSM)。Master-MSM被证明适用于广泛的作用力条件。我们证明了该理论在三种不同生物分子系统中的应用,即十丙氨酸,TBA(凝血酶结合适体)和RNA发夹。
更新日期:2018-09-13
中文翻译:

操纵分子动力学加速构建生物分子动力学网络模型
介绍了基于转向分子动力学(SMD)模拟的一类新的罕见事件加速技术。施加在生物分子上的拉伸力使其进入较大的端到端距离。在这些条件下,生物分子会经历快速构象变化,这在零力条件下是罕见的。提出了描述生物分子在各种拉伸力下的动力学的理论。使用该理论,可以从在很小的力条件范围内经常访问的速率构造主-马尔可夫状态模型(master-MSM)。Master-MSM被证明适用于广泛的作用力条件。我们证明了该理论在三种不同生物分子系统中的应用,即十丙氨酸,TBA(凝血酶结合适体)和RNA发夹。



























 京公网安备 11010802027423号
京公网安备 11010802027423号