Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
De novo design of a fluorescence-activating β-barrel
Nature ( IF 64.8 ) Pub Date : 2018-09-01 , DOI: 10.1038/s41586-018-0509-0 Jiayi Dou 1, 2 , Anastassia A Vorobieva 1, 2 , William Sheffler 1, 2 , Lindsey A Doyle 3 , Hahnbeom Park 1, 2 , Matthew J Bick 1, 2 , Binchen Mao 1, 4 , Glenna W Foight 5 , Min Yen Lee 5 , Lauren A Gagnon 5 , Lauren Carter 1, 2 , Banumathi Sankaran 6 , Sergey Ovchinnikov 1, 2, 7 , Enrique Marcos 1, 2, 8 , Po-Ssu Huang 1, 2, 9 , Joshua C Vaughan 5 , Barry L Stoddard 3 , David Baker 1, 2, 10
Nature ( IF 64.8 ) Pub Date : 2018-09-01 , DOI: 10.1038/s41586-018-0509-0 Jiayi Dou 1, 2 , Anastassia A Vorobieva 1, 2 , William Sheffler 1, 2 , Lindsey A Doyle 3 , Hahnbeom Park 1, 2 , Matthew J Bick 1, 2 , Binchen Mao 1, 4 , Glenna W Foight 5 , Min Yen Lee 5 , Lauren A Gagnon 5 , Lauren Carter 1, 2 , Banumathi Sankaran 6 , Sergey Ovchinnikov 1, 2, 7 , Enrique Marcos 1, 2, 8 , Po-Ssu Huang 1, 2, 9 , Joshua C Vaughan 5 , Barry L Stoddard 3 , David Baker 1, 2, 10
Affiliation
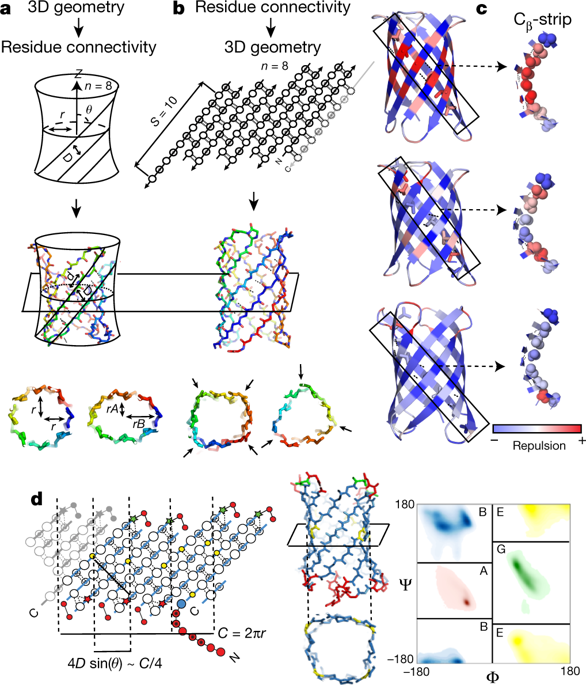
|
The regular arrangements of β-strands around a central axis in β-barrels and of α-helices in coiled coils contrast with the irregular tertiary structures of most globular proteins, and have fascinated structural biologists since they were first discovered. Simple parametric models have been used to design a wide range of α-helical coiled-coil structures, but to date there has been no success with β-barrels. Here we show that accurate de novo design of β-barrels requires considerable symmetry-breaking to achieve continuous hydrogen-bond connectivity and eliminate backbone strain. We then build ensembles of β-barrel backbone models with cavity shapes that match the fluorogenic compound DFHBI, and use a hierarchical grid-based search method to simultaneously optimize the rigid-body placement of DFHBI in these cavities and the identities of the surrounding amino acids to achieve high shape and chemical complementarity. The designs have high structural accuracy and bind and fluorescently activate DFHBI in vitro and in Escherichia coli, yeast and mammalian cells. This de novo design of small-molecule binding activity, using backbones custom-built to bind the ligand, should enable the design of increasingly sophisticated ligand-binding proteins, sensors and catalysts that are not limited by the backbone geometries available in known protein structures.The elucidation of general principles for designing β-barrels enables the de novo creation of fluorescent proteins.
中文翻译:

荧光激活β-桶的从头设计
β 链在 β 桶中围绕中心轴的规则排列和 α 螺旋在盘绕螺旋中的规则排列与大多数球状蛋白质的不规则三级结构形成对比,并且自从它们首次被发现以来就让结构生物学家着迷。简单的参数模型已用于设计范围广泛的 α 螺旋卷曲螺旋结构,但迄今为止,β 桶尚未取得成功。在这里,我们表明 β 桶的准确从头设计需要相当大的对称性破坏才能实现连续的氢键连接并消除骨架应变。然后,我们构建了具有与荧光化合物 DFHBI 相匹配的空腔形状的 β 桶骨干模型的集合,并使用基于分层网格的搜索方法同时优化 DFHBI 在这些空腔中的刚体放置和周围氨基酸的特性,以实现高度的形状和化学互补性。这些设计具有高结构准确性,并在体外和大肠杆菌、酵母和哺乳动物细胞中结合并荧光激活 DFHBI。这种小分子结合活性的从头设计,使用定制的主链来结合配体,应该能够设计出越来越复杂的配体结合蛋白、传感器和催化剂,这些蛋白、传感器和催化剂不受已知蛋白质结构中可用的主链几何形状的限制。阐明设计 β 桶的一般原则可以从头创建荧光蛋白。这些设计具有高结构准确性,并在体外和大肠杆菌、酵母和哺乳动物细胞中结合并荧光激活 DFHBI。这种小分子结合活性的从头设计,使用定制的主链来结合配体,应该能够设计出越来越复杂的配体结合蛋白、传感器和催化剂,这些蛋白、传感器和催化剂不受已知蛋白质结构中可用的主链几何形状的限制。阐明设计 β 桶的一般原则可以从头创建荧光蛋白。这些设计具有高结构准确性,并在体外和大肠杆菌、酵母和哺乳动物细胞中结合并荧光激活 DFHBI。这种小分子结合活性的从头设计,使用定制的主链来结合配体,应该能够设计出越来越复杂的配体结合蛋白、传感器和催化剂,这些蛋白、传感器和催化剂不受已知蛋白质结构中可用的主链几何形状的限制。阐明设计 β 桶的一般原则可以从头创建荧光蛋白。
更新日期:2018-09-01
中文翻译:

荧光激活β-桶的从头设计
β 链在 β 桶中围绕中心轴的规则排列和 α 螺旋在盘绕螺旋中的规则排列与大多数球状蛋白质的不规则三级结构形成对比,并且自从它们首次被发现以来就让结构生物学家着迷。简单的参数模型已用于设计范围广泛的 α 螺旋卷曲螺旋结构,但迄今为止,β 桶尚未取得成功。在这里,我们表明 β 桶的准确从头设计需要相当大的对称性破坏才能实现连续的氢键连接并消除骨架应变。然后,我们构建了具有与荧光化合物 DFHBI 相匹配的空腔形状的 β 桶骨干模型的集合,并使用基于分层网格的搜索方法同时优化 DFHBI 在这些空腔中的刚体放置和周围氨基酸的特性,以实现高度的形状和化学互补性。这些设计具有高结构准确性,并在体外和大肠杆菌、酵母和哺乳动物细胞中结合并荧光激活 DFHBI。这种小分子结合活性的从头设计,使用定制的主链来结合配体,应该能够设计出越来越复杂的配体结合蛋白、传感器和催化剂,这些蛋白、传感器和催化剂不受已知蛋白质结构中可用的主链几何形状的限制。阐明设计 β 桶的一般原则可以从头创建荧光蛋白。这些设计具有高结构准确性,并在体外和大肠杆菌、酵母和哺乳动物细胞中结合并荧光激活 DFHBI。这种小分子结合活性的从头设计,使用定制的主链来结合配体,应该能够设计出越来越复杂的配体结合蛋白、传感器和催化剂,这些蛋白、传感器和催化剂不受已知蛋白质结构中可用的主链几何形状的限制。阐明设计 β 桶的一般原则可以从头创建荧光蛋白。这些设计具有高结构准确性,并在体外和大肠杆菌、酵母和哺乳动物细胞中结合并荧光激活 DFHBI。这种小分子结合活性的从头设计,使用定制的主链来结合配体,应该能够设计出越来越复杂的配体结合蛋白、传感器和催化剂,这些蛋白、传感器和催化剂不受已知蛋白质结构中可用的主链几何形状的限制。阐明设计 β 桶的一般原则可以从头创建荧光蛋白。



























 京公网安备 11010802027423号
京公网安备 11010802027423号