当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structural Dynamics of Lateral and Diagonal Loops of Human Telomeric G-Quadruplexes in Extended MD Simulations
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-09-05 00:00:00 , DOI: 10.1021/acs.jctc.8b00543 Barira Islam 1 , Petr Stadlbauer 1 , Miroslav Krepl 1 , Marek Havrila 1 , Shozeb Haider 2 , Jiri Sponer 1
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-09-05 00:00:00 , DOI: 10.1021/acs.jctc.8b00543 Barira Islam 1 , Petr Stadlbauer 1 , Miroslav Krepl 1 , Marek Havrila 1 , Shozeb Haider 2 , Jiri Sponer 1
Affiliation
|
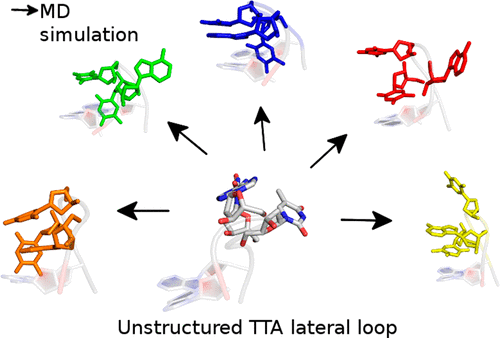
The NMR solution structures of human telomeric (Htel) G-quadruplexes (GQs) are characterized by the presence of two lateral loops complemented by either diagonal or propeller loops. Bases of a given loop can establish interactions within the loop as well as with other loops and the flanking bases. This can lead to a formation of base alignments above and below the GQ stems. These base alignments are known to affect the loop structures and relative stabilities of different Htel GQ folds. We have carried out a total of 217 μs of classical (unbiased) molecular dynamics (MD) simulations starting from the available solution structures of Htel GQs to characterize structural dynamics of the lateral and diagonal loops, using several recent AMBER DNA force-field variants. As the loops are involved in diverse stacking and H-bonding interactions, their dynamics is slow, and extended sampling is required to capture different conformations. Nevertheless, although the simulations are far from being quantitatively converged, the data suggest that multiple 10 μs-scale simulations can provide a quite good assessment of the loop conformational space as described by the force field. The simulations indicate that the lateral loops may sample multiple coexisting conformations, which should be considered when comparing simulations with the NMR models as the latter include ensemble averaging. The adenine–thymine Watson–Crick arrangement was the most stable base pairing in the simulations. Adenine–adenine and thymine–thymine base pairs were also sampled but were less stable. The data suggest that the description of lateral and diagonal GQ loops in contemporary MD simulations is considerably more realistic than the description of propeller loops, though definitely not flawless.
中文翻译:

人类端粒G四联体的横向和对角线环的结构动力学在扩展的MD模拟中
人类端粒(Htel)G-四链体(GQs)的NMR溶液结构的特征是存在两个侧向环,对角线或螺旋桨环互补。给定循环的基数可以在该循环内以及与其他循环和侧基之间建立相互作用。这可能导致在GQ茎上方和下方形成碱基比对。已知这些碱基比对影响不同Htel GQ折叠的环结构和相对稳定性。我们从Htel GQ的可用溶液结构开始,总共进行了217μs的经典(无偏)分子动力学(MD)模拟,以使用几个最新的AMBER DNA力场变体表征侧向和对角环的结构动力学。由于循环涉及各种堆叠和氢键相互作用,它们的动力学很慢,需要扩展采样来捕获不同的构象。尽管如此,尽管仿真还远未达到定量收敛的目的,但数据表明,多个10μs规模的仿真可以对力场所描述的环构象空间进行很好的评估。仿真表明,横向环可能会采样多个共存的构象,将仿真与NMR模型进行比较时应考虑这些因素,因为后者包括整体平均。腺嘌呤-胸腺嘧啶沃森-克里克安排是模拟中最稳定的碱基配对。还对腺嘌呤-腺嘌呤和胸腺嘧啶-胸腺嘧啶碱基对进行了采样,但稳定性较差。
更新日期:2018-09-05
中文翻译:

人类端粒G四联体的横向和对角线环的结构动力学在扩展的MD模拟中
人类端粒(Htel)G-四链体(GQs)的NMR溶液结构的特征是存在两个侧向环,对角线或螺旋桨环互补。给定循环的基数可以在该循环内以及与其他循环和侧基之间建立相互作用。这可能导致在GQ茎上方和下方形成碱基比对。已知这些碱基比对影响不同Htel GQ折叠的环结构和相对稳定性。我们从Htel GQ的可用溶液结构开始,总共进行了217μs的经典(无偏)分子动力学(MD)模拟,以使用几个最新的AMBER DNA力场变体表征侧向和对角环的结构动力学。由于循环涉及各种堆叠和氢键相互作用,它们的动力学很慢,需要扩展采样来捕获不同的构象。尽管如此,尽管仿真还远未达到定量收敛的目的,但数据表明,多个10μs规模的仿真可以对力场所描述的环构象空间进行很好的评估。仿真表明,横向环可能会采样多个共存的构象,将仿真与NMR模型进行比较时应考虑这些因素,因为后者包括整体平均。腺嘌呤-胸腺嘧啶沃森-克里克安排是模拟中最稳定的碱基配对。还对腺嘌呤-腺嘌呤和胸腺嘧啶-胸腺嘧啶碱基对进行了采样,但稳定性较差。



























 京公网安备 11010802027423号
京公网安备 11010802027423号