Bioorganic & Medicinal Chemistry Letters ( IF 2.7 ) Pub Date : 2018-08-27 , DOI: 10.1016/j.bmcl.2018.08.032 Yoshihiro Uesawa
|
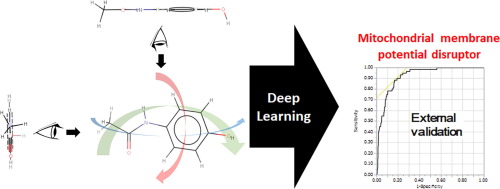
Quantitative structure–activity relationship (QSAR) analysis uses structural, quantum chemical, and physicochemical features calculated from molecular geometry as explanatory variables predicting physiological activity. Recently, deep learning based on advanced artificial neural networks has demonstrated excellent performance in the discipline of QSAR research. While it has properties of feature representation learning that directly calculate feature values from molecular structure, the use of this potential function is limited in QSAR modeling. The present study applied this function of feature representation learning to QSAR analysis by incorporating 360° images of molecular conformations into deep learning. Accordingly, I successfully constructed a highly versatile identification model for chemical compounds that induce mitochondrial membrane potential disruption with the external validation area under the receiver operating characteristic curve of ≥0.9.
中文翻译:

基于新型分子图像输入技术的深度学习定量构效关系分析
定量构效关系(QSAR)分析使用从分子几何学计算得到的结构,量子化学和物理化学特征作为预测生理活性的解释变量。近年来,基于高级人工神经网络的深度学习已在QSAR研究领域表现出出色的性能。虽然它具有直接从分子结构计算特征值的特征表示学习的特性,但在QSAR建模中,此潜在功能的使用受到限制。本研究通过将分子构象的360°图像整合到深度学习中,将特征表示学习的功能应用于QSAR分析。因此,


























 京公网安备 11010802027423号
京公网安备 11010802027423号