当前位置:
X-MOL 学术
›
Macromol. Rapid Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Synthesis of Single‐Ring Nanoparticles Mimicking Natural Cyclotides by a Stepwise Folding‐Activation‐Collapse Process
Macromolecular Rapid Communications ( IF 4.6 ) Pub Date : 2018-08-24 , DOI: 10.1002/marc.201800491 Jon Rubio-Cervilla 1 , Hendrik Frisch 2 , Christopher Barner-Kowollik 2, 3 , José A. Pomposo 1, 4, 5
Macromolecular Rapid Communications ( IF 4.6 ) Pub Date : 2018-08-24 , DOI: 10.1002/marc.201800491 Jon Rubio-Cervilla 1 , Hendrik Frisch 2 , Christopher Barner-Kowollik 2, 3 , José A. Pomposo 1, 4, 5
Affiliation
|
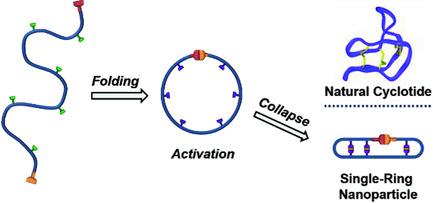
Cyclotides are small cyclic polypeptides found in a variety of organisms, ranging from bacteria to plants. Their ring structure endows those polypeptides with specific properties, such as improved stability against enzymatic degradation. Optimal cyclotide activity is often observed only in the presence of intra‐ring disulfide bonds. Synthesis of soft nano‐objects mimicking the conformation of natural cyclotides remains challenging. Here, a new class of natural cyclotide mimics synthesized by a stepwise folding‐activation‐collapse process at high dilution starting from simple synthetic precursor polymers is established. The initial folding step is carried out by a photoactivated hetero Diels–Alder (HDA) ring‐closing reaction, which is accompanied by chain compaction of the individual precursor polymer chains as determined by size exclusion chromatography (SEC). The subsequent activation step comprises a simple azidation procedure, whereas the final collapse step is driven by CuAAC in the presence of an external cross‐linker, providing additional compaction to the final single‐ring nanoparticles (SRNPs). The unique structure and compaction degree of the SRNPs is established via a detailed comparison with conventional single‐chain nanoparticles (SCNPs) prepared exclusively by chain collapse from the exact same precursor polymer (without the prefolding step). The stepwise folding‐activation‐collapse approach opens new avenues for the preparation of artificial cyclotide mimetics.
中文翻译:

通过逐步折叠-活化-折叠过程合成模仿天然环氧化物的单环纳米颗粒
环肽是存在于多种生物中的小环状多肽,从细菌到植物。它们的环结构赋予了这些多肽特定的特性,例如提高了对酶促降解的稳定性。通常仅在存在环内二硫键的情况下才能观察到最佳的环氧化物活性。模仿天然环氧化物构象的软纳米物体的合成仍然具有挑战性。在此,从简单的合成前体聚合物开始,通过逐步的折叠-活化-折叠过程以高稀释度合成了一类新的天然环氧化物模拟物。初始折叠步骤是通过光活化杂Diels-Alder(HDA)闭环反应进行的,如尺寸排阻色谱法(SEC)所述,其伴随着单个前体聚合物链的链压缩。随后的激活步骤包括一个简单的叠氮化步骤,而最终的折叠步骤是在外部交联剂存在下由CuAAC驱动的,从而为最终的单环纳米颗粒(SRNP)提供了额外的压实性。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。而最终的塌缩步骤是在外部交联剂存在下由CuAAC驱动的,从而为最终的单环纳米颗粒(SRNP)提供了额外的压实性。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。而最终的塌缩步骤是在外部交联剂存在下由CuAAC驱动的,从而为最终的单环纳米颗粒(SRNP)提供了额外的压实性。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。
更新日期:2018-08-24
中文翻译:

通过逐步折叠-活化-折叠过程合成模仿天然环氧化物的单环纳米颗粒
环肽是存在于多种生物中的小环状多肽,从细菌到植物。它们的环结构赋予了这些多肽特定的特性,例如提高了对酶促降解的稳定性。通常仅在存在环内二硫键的情况下才能观察到最佳的环氧化物活性。模仿天然环氧化物构象的软纳米物体的合成仍然具有挑战性。在此,从简单的合成前体聚合物开始,通过逐步的折叠-活化-折叠过程以高稀释度合成了一类新的天然环氧化物模拟物。初始折叠步骤是通过光活化杂Diels-Alder(HDA)闭环反应进行的,如尺寸排阻色谱法(SEC)所述,其伴随着单个前体聚合物链的链压缩。随后的激活步骤包括一个简单的叠氮化步骤,而最终的折叠步骤是在外部交联剂存在下由CuAAC驱动的,从而为最终的单环纳米颗粒(SRNP)提供了额外的压实性。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。而最终的塌缩步骤是在外部交联剂存在下由CuAAC驱动的,从而为最终的单环纳米颗粒(SRNP)提供了额外的压实性。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。而最终的塌缩步骤是在外部交联剂存在下由CuAAC驱动的,从而为最终的单环纳米颗粒(SRNP)提供了额外的压实性。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。SRNP的独特结构和紧密度是通过与传统单链纳米颗粒(SCNP)的详细比较而确定的,该常规单链纳米颗粒仅由完全相同的前体聚合物进行链折叠而制得(无需预折叠步骤)。逐步折叠-活化-折叠方法为制备人工环氧化物模拟物开辟了新途径。



























 京公网安备 11010802027423号
京公网安备 11010802027423号