当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Robust and Quantitative Reporter System To Evaluate Noncanonical Amino Acid Incorporation in Yeast
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-08-23 00:00:00 , DOI: 10.1021/acssynbio.8b00260 Jessica T Stieglitz 1 , Haixing P Kehoe 1 , Ming Lei 1 , James A Van Deventer 1, 2
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-08-23 00:00:00 , DOI: 10.1021/acssynbio.8b00260 Jessica T Stieglitz 1 , Haixing P Kehoe 1 , Ming Lei 1 , James A Van Deventer 1, 2
Affiliation
|
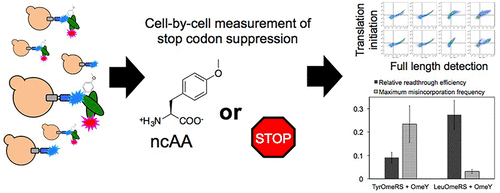
Engineering protein translation machinery to incorporate noncanonical amino acids (ncAAs) into proteins has advanced applications ranging from proteomics to single-molecule studies. As applications of ncAAs emerge, efficient ncAA incorporation is crucial to exploiting unique chemistries. We have established a quantitative reporter platform to evaluate ncAA incorporation in response to the TAG (amber) codon in yeast. This yeast display-based reporter utilizes an antibody fragment containing an amber codon at which a ncAA is incorporated when the appropriate orthogonal translation system (OTS) is present. Epitope tags at both termini allow for flow cytometry-based end point readouts of OTS efficiency and fidelity. Using this reporter, we evaluated several factors that influence amber suppression, including the amber codon position and different aminoacyl-tRNA synthetase/tRNA (aaRS/tRNA) pairs. Interestingly, previously described aaRSs that evolved from different parent enzymes to incorporate O-methyl-l-tyrosine exhibit vastly different behavior. Escherichia coli leucyl-tRNA synthetase variants demonstrated efficient incorporation of a range of ncAAs, and we discovered unreported activities of several variants. Compared to a plate reader-based reporter, our assay yields more precise bulk-level measurements while also supporting single-cell readouts compatible with cell sorting. This platform is expected to allow quantitative elucidation of principles dictating efficient stop codon suppression and evolution of next-generation stop codon suppression systems to further enhance genetic code manipulation in eukaryotes. These efforts will improve our understanding of how the genetic code can be further evolved while expanding the range of chemical diversity available in proteins for applications ranging from fundamental epigenetics studies to engineering new classes of therapeutics.
中文翻译:

用于评估酵母中非规范氨基酸掺入的稳健定量报告系统
工程蛋白质翻译机器将非规范氨基酸 (ncAA) 整合到蛋白质中,具有从蛋白质组学到单分子研究的先进应用。随着 ncAA 应用的出现,有效的 ncAA 掺入对于开发独特的化学物质至关重要。我们建立了一个定量报告平台来评估 ncAA 掺入对酵母中 TAG(琥珀)密码子的反应。这种基于酵母展示的报告基因利用含有琥珀密码子的抗体片段,当存在适当的正交翻译系统 (OTS) 时,该密码子中会掺入 ncAA。两个末端的表位标签允许基于流式细胞术的 OTS 效率和保真度终点读数。使用该报告基因,我们评估了影响琥珀抑制的几个因素,包括琥珀密码子位置和不同的氨酰基-tRNA 合成酶/tRNA (aaRS/tRNA) 对。有趣的是,之前描述的从不同亲本酶进化而来并入O-甲基-L-酪氨酸的aaRS表现出截然不同的行为。大肠杆菌亮氨酰-tRNA 合成酶变体证明了一系列 ncAA 的有效掺入,并且我们发现了几种未报道的变体活性。与基于酶标仪的报告仪相比,我们的测定可产生更精确的批量水平测量,同时还支持与细胞分选兼容的单细胞读数。该平台有望定量阐明有效终止密码子抑制的原理以及下一代终止密码子抑制系统的进化,以进一步增强真核生物中的遗传密码操作。这些努力将增进我们对遗传密码如何进一步进化的理解,同时扩大蛋白质中可用的化学多样性范围,适用于从基础表观遗传学研究到工程新类别治疗的应用。
更新日期:2018-08-23
中文翻译:

用于评估酵母中非规范氨基酸掺入的稳健定量报告系统
工程蛋白质翻译机器将非规范氨基酸 (ncAA) 整合到蛋白质中,具有从蛋白质组学到单分子研究的先进应用。随着 ncAA 应用的出现,有效的 ncAA 掺入对于开发独特的化学物质至关重要。我们建立了一个定量报告平台来评估 ncAA 掺入对酵母中 TAG(琥珀)密码子的反应。这种基于酵母展示的报告基因利用含有琥珀密码子的抗体片段,当存在适当的正交翻译系统 (OTS) 时,该密码子中会掺入 ncAA。两个末端的表位标签允许基于流式细胞术的 OTS 效率和保真度终点读数。使用该报告基因,我们评估了影响琥珀抑制的几个因素,包括琥珀密码子位置和不同的氨酰基-tRNA 合成酶/tRNA (aaRS/tRNA) 对。有趣的是,之前描述的从不同亲本酶进化而来并入O-甲基-L-酪氨酸的aaRS表现出截然不同的行为。大肠杆菌亮氨酰-tRNA 合成酶变体证明了一系列 ncAA 的有效掺入,并且我们发现了几种未报道的变体活性。与基于酶标仪的报告仪相比,我们的测定可产生更精确的批量水平测量,同时还支持与细胞分选兼容的单细胞读数。该平台有望定量阐明有效终止密码子抑制的原理以及下一代终止密码子抑制系统的进化,以进一步增强真核生物中的遗传密码操作。这些努力将增进我们对遗传密码如何进一步进化的理解,同时扩大蛋白质中可用的化学多样性范围,适用于从基础表观遗传学研究到工程新类别治疗的应用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号