当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Evaluation of APOBEC3B Recognition Motifs by NMR Reveals Preferred Substrates.
ACS Chemical Biology ( IF 4 ) Pub Date : 2018-08-27 , DOI: 10.1021/acschembio.8b00639 Manjuan Liu 1 , Aurélie Mallinger 1 , Marcello Tortorici 1 , Yvette Newbatt 1 , Meirion Richards 1 , Amin Mirza 1 , Rob L M van Montfort 1 , Rosemary Burke 1 , Julian Blagg 1 , Teresa Kaserer 1
ACS Chemical Biology ( IF 4 ) Pub Date : 2018-08-27 , DOI: 10.1021/acschembio.8b00639 Manjuan Liu 1 , Aurélie Mallinger 1 , Marcello Tortorici 1 , Yvette Newbatt 1 , Meirion Richards 1 , Amin Mirza 1 , Rob L M van Montfort 1 , Rosemary Burke 1 , Julian Blagg 1 , Teresa Kaserer 1
Affiliation
|
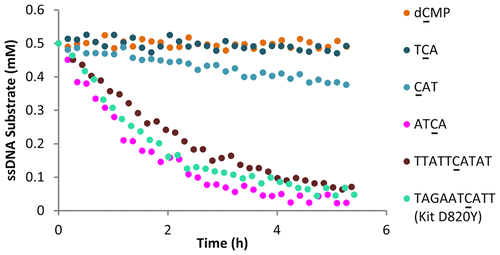
APOBEC3B (A3B) deamination activity on ssDNA is considered a contributing factor to tumor heterogeneity and drug resistance in a number of human cancers. Despite its clinical impact, little is known about A3B ssDNA substrate preference. We have used nuclear magnetic resonance to monitor the catalytic turnover of A3B substrates in real-time. This study reports preferred nucleotide sequences for A3B substrates, including optimized 4-mer oligonucleotides, and reveals a breadth of substrate recognition that includes DNA sequences known to be mutated in drug-resistant cancer clones. Our results are consistent with available clinical and structural data and may inform the design of substrate-based A3B inhibitors.
中文翻译:

核磁共振对 APOBEC3B 识别基序的评估揭示了优选的底物。
ssDNA 上的 APOBEC3B (A3B) 脱氨基活性被认为是许多人类癌症中肿瘤异质性和耐药性的一个促成因素。尽管具有临床影响,但对 A3B ssDNA 底物偏好知之甚少。我们使用核磁共振实时监测 A3B 底物的催化转换。该研究报告了 A3B 底物的首选核苷酸序列,包括优化的 4 聚体寡核苷酸,并揭示了广泛的底物识别,包括已知在耐药癌症克隆中发生突变的 DNA 序列。我们的结果与现有的临床和结构数据一致,并可能为基于底物的 A3B 抑制剂的设计提供信息。
更新日期:2018-08-21
中文翻译:

核磁共振对 APOBEC3B 识别基序的评估揭示了优选的底物。
ssDNA 上的 APOBEC3B (A3B) 脱氨基活性被认为是许多人类癌症中肿瘤异质性和耐药性的一个促成因素。尽管具有临床影响,但对 A3B ssDNA 底物偏好知之甚少。我们使用核磁共振实时监测 A3B 底物的催化转换。该研究报告了 A3B 底物的首选核苷酸序列,包括优化的 4 聚体寡核苷酸,并揭示了广泛的底物识别,包括已知在耐药癌症克隆中发生突变的 DNA 序列。我们的结果与现有的临床和结构数据一致,并可能为基于底物的 A3B 抑制剂的设计提供信息。


























 京公网安备 11010802027423号
京公网安备 11010802027423号