当前位置:
X-MOL 学术
›
ChemMedChem
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structural Basis of Substrate Recognition and Covalent Inhibition of Cdu1 from Chlamydia trachomatis
ChemMedChem ( IF 3.4 ) Pub Date : 2018-09-06 , DOI: 10.1002/cmdc.201800364 Yesid A Ramirez 1, 2 , Thomas B Adler 2 , Eva Altmann 3 , Theresa Klemm 1 , Christian Tiesmeyer 1 , Florian Sauer 1 , Stefan G Kathman 4 , Alexander V Statsyuk 5 , Christoph Sotriffer 2 , Caroline Kisker 1
ChemMedChem ( IF 3.4 ) Pub Date : 2018-09-06 , DOI: 10.1002/cmdc.201800364 Yesid A Ramirez 1, 2 , Thomas B Adler 2 , Eva Altmann 3 , Theresa Klemm 1 , Christian Tiesmeyer 1 , Florian Sauer 1 , Stefan G Kathman 4 , Alexander V Statsyuk 5 , Christoph Sotriffer 2 , Caroline Kisker 1
Affiliation
|
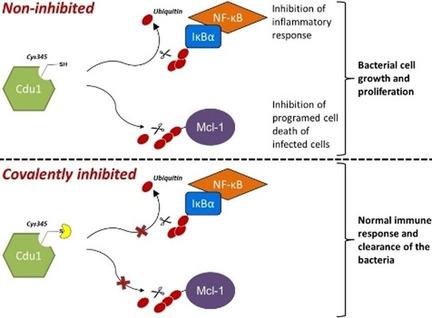
Based on the similarity between the active sites of the deubiquitylating and deneddylating enzyme ChlaDub1 (Cdu1) and the evolutionarily related protease adenain, a target‐hopping screening approach on a focused set of adenain inhibitors was investigated. The cyanopyrimidine‐based inhibitors identified represent the first active‐site‐directed small‐molecule inhibitors of Cdu1. High‐resolution crystal structures of Cdu1 in complex with two covalently bound cyanopyrimidines, as well as with its substrate ubiquitin, were obtained. These structural data were complemented by enzymatic assays and covalent docking studies to provide insight into the substrate recognition of Cdu1, active‐site pocket flexibility and potential hotspots for ligand interaction. Combined, these data provide a strong basis for future structure‐guided medicinal chemistry optimization of this cyanopyrimidine scaffold into more potent and selective Cdu1 inhibitors.
中文翻译:

沙眼衣原体 Cdu1 的底物识别和共价抑制的结构基础
基于去泛素化和去甲基化酶 ChlaDub1 (Cdu1) 的活性位点与进化相关的蛋白酶 adenin 之间的相似性,研究了针对一组集中的 adenin 抑制剂的靶标跳跃筛选方法。确定的基于氰基嘧啶的抑制剂代表了第一个活性位点定向的 Cdu1 小分子抑制剂。获得了 Cdu1 与两个共价结合的氰基嘧啶及其底物泛素复合物的高分辨率晶体结构。这些结构数据通过酶测定和共价对接研究得到补充,以深入了解 Cdu1 的底物识别、活性位点口袋灵活性和配体相互作用的潜在热点。综合起来,这些数据为未来将氰基嘧啶支架结构引导的药物化学优化为更有效、更具选择性的 Cdu1 抑制剂提供了坚实的基础。
更新日期:2018-09-06
中文翻译:

沙眼衣原体 Cdu1 的底物识别和共价抑制的结构基础
基于去泛素化和去甲基化酶 ChlaDub1 (Cdu1) 的活性位点与进化相关的蛋白酶 adenin 之间的相似性,研究了针对一组集中的 adenin 抑制剂的靶标跳跃筛选方法。确定的基于氰基嘧啶的抑制剂代表了第一个活性位点定向的 Cdu1 小分子抑制剂。获得了 Cdu1 与两个共价结合的氰基嘧啶及其底物泛素复合物的高分辨率晶体结构。这些结构数据通过酶测定和共价对接研究得到补充,以深入了解 Cdu1 的底物识别、活性位点口袋灵活性和配体相互作用的潜在热点。综合起来,这些数据为未来将氰基嘧啶支架结构引导的药物化学优化为更有效、更具选择性的 Cdu1 抑制剂提供了坚实的基础。



























 京公网安备 11010802027423号
京公网安备 11010802027423号