Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Complex silica composite nanomaterials templated with DNA origami
Nature ( IF 64.8 ) Pub Date : 2018-07-01 , DOI: 10.1038/s41586-018-0332-7 Xiaoguo Liu 1, 2 , Fei Zhang 3, 4 , Xinxin Jing 1 , Muchen Pan 1 , Pi Liu 5, 6 , Wei Li 7 , Bowen Zhu 1 , Jiang Li 1, 8 , Hong Chen 9 , Lihua Wang 1 , Jianping Lin 5, 6 , Yan Liu 3, 4 , Dongyuan Zhao 7 , Hao Yan 3, 4 , Chunhai Fan 1
Nature ( IF 64.8 ) Pub Date : 2018-07-01 , DOI: 10.1038/s41586-018-0332-7 Xiaoguo Liu 1, 2 , Fei Zhang 3, 4 , Xinxin Jing 1 , Muchen Pan 1 , Pi Liu 5, 6 , Wei Li 7 , Bowen Zhu 1 , Jiang Li 1, 8 , Hong Chen 9 , Lihua Wang 1 , Jianping Lin 5, 6 , Yan Liu 3, 4 , Dongyuan Zhao 7 , Hao Yan 3, 4 , Chunhai Fan 1
Affiliation
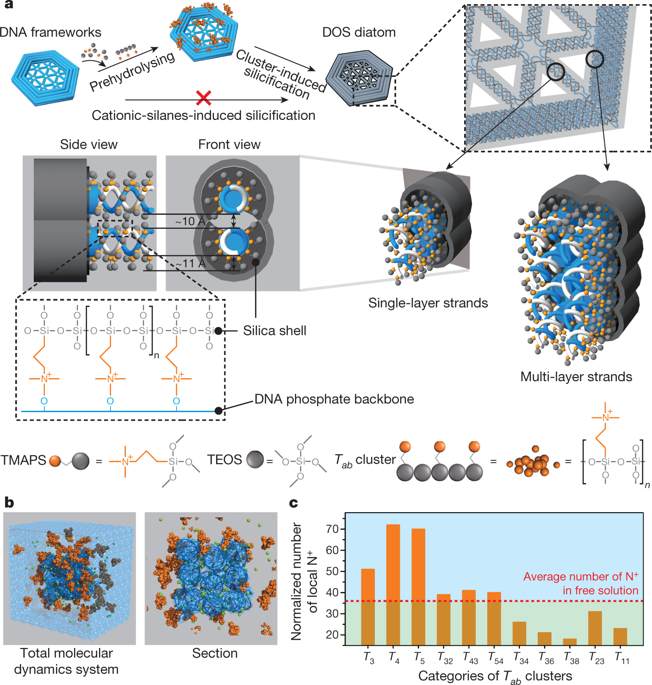
|
Genetically encoded protein scaffolds often serve as templates for the mineralization of biocomposite materials with complex yet highly controlled structural features that span from nanometres to the macroscopic scale1–4. Methods developed to mimic these fabrication capabilities can produce synthetic materials with well defined micro- and macro-sized features, but extending control to the nanoscale remains challenging5,6. DNA nanotechnology can deliver a wide range of customized nanoscale two- and three-dimensional assemblies with controlled sizes and shapes7–11. But although DNA has been used to modulate the morphology of inorganic materials12,13 and DNA nanostructures have served as moulds14,15 and templates16,17, it remains challenging to exploit the potential of DNA nanostructures fully because they require high-ionic-strength solutions to maintain their structure, and this in turn gives rise to surface charging that suppresses the material deposition. Here we report that the Stöber method, widely used for producing silica (silicon dioxide) nanostructures, can be adjusted to overcome this difficulty: when synthesis conditions are such that mineral precursor molecules do not deposit directly but first form clusters, DNA–silica hybrid materials that faithfully replicate the complex geometric information of a wide range of different DNA origami scaffolds are readily obtained. We illustrate this approach using frame-like, curved and porous DNA nanostructures, with one-, two- and three-dimensional complex hierarchical architectures that range in size from 10 to 1,000 nanometres. We also show that after coating with an amorphous silica layer, the thickness of which can be tuned by adjusting the growth time, hybrid structures can be up to ten times tougher than the DNA template while maintaining flexibility. These findings establish our approach as a general method for creating biomimetic silica nanostructures.DNA origami is used as a template to produce complex geometric shapes of nanoscale silica hybrid materials.
中文翻译:

以DNA折纸为模板的复杂二氧化硅复合纳米材料
基因编码的蛋白质支架通常作为生物复合材料矿化的模板,具有复杂但高度可控的结构特征,从纳米到宏观尺度1-4。为模仿这些制造能力而开发的方法可以生产具有明确定义的微观和宏观尺寸特征的合成材料,但将控制扩展到纳米尺度仍然具有挑战性5,6。DNA 纳米技术可以提供各种尺寸和形状可控的定制纳米级二维和三维组件7-11。但是,尽管 DNA 已被用于调节无机材料的形态 12,13,并且 DNA 纳米结构已用作模具 14,15 和模板 16,17,充分利用 DNA 纳米结构的潜力仍然具有挑战性,因为它们需要高离子强度的溶液来维持其结构,而这反过来又会产生抑制材料沉积的表面电荷。在这里,我们报告了广泛用于生产二氧化硅(二氧化硅)纳米结构的 Stöber 方法可以调整以克服这一困难:当合成条件使得矿物前体分子不直接沉积而是首先形成簇时,DNA-二氧化硅杂化材料忠实地复制各种不同 DNA 折纸支架的复杂几何信息是很容易获得的。我们使用框架状、弯曲和多孔 DNA 纳米结构来说明这种方法,其中一个,二维和三维复杂的层次结构,尺寸范围从 10 到 1,000 纳米。我们还表明,在涂有无定形二氧化硅层后,其厚度可以通过调整生长时间来调整,混合结构的韧性可以比 DNA 模板高 10 倍,同时保持柔韧性。这些发现确立了我们作为创建仿生二氧化硅纳米结构的一般方法的方法。DNA折纸被用作模板来生产纳米级二氧化硅杂化材料的复杂几何形状。
更新日期:2018-07-01
中文翻译:

以DNA折纸为模板的复杂二氧化硅复合纳米材料
基因编码的蛋白质支架通常作为生物复合材料矿化的模板,具有复杂但高度可控的结构特征,从纳米到宏观尺度1-4。为模仿这些制造能力而开发的方法可以生产具有明确定义的微观和宏观尺寸特征的合成材料,但将控制扩展到纳米尺度仍然具有挑战性5,6。DNA 纳米技术可以提供各种尺寸和形状可控的定制纳米级二维和三维组件7-11。但是,尽管 DNA 已被用于调节无机材料的形态 12,13,并且 DNA 纳米结构已用作模具 14,15 和模板 16,17,充分利用 DNA 纳米结构的潜力仍然具有挑战性,因为它们需要高离子强度的溶液来维持其结构,而这反过来又会产生抑制材料沉积的表面电荷。在这里,我们报告了广泛用于生产二氧化硅(二氧化硅)纳米结构的 Stöber 方法可以调整以克服这一困难:当合成条件使得矿物前体分子不直接沉积而是首先形成簇时,DNA-二氧化硅杂化材料忠实地复制各种不同 DNA 折纸支架的复杂几何信息是很容易获得的。我们使用框架状、弯曲和多孔 DNA 纳米结构来说明这种方法,其中一个,二维和三维复杂的层次结构,尺寸范围从 10 到 1,000 纳米。我们还表明,在涂有无定形二氧化硅层后,其厚度可以通过调整生长时间来调整,混合结构的韧性可以比 DNA 模板高 10 倍,同时保持柔韧性。这些发现确立了我们作为创建仿生二氧化硅纳米结构的一般方法的方法。DNA折纸被用作模板来生产纳米级二氧化硅杂化材料的复杂几何形状。



























 京公网安备 11010802027423号
京公网安备 11010802027423号