当前位置:
X-MOL 学术
›
Phys. Chem. Chem. Phys.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Simulated and experimental force spectroscopy of lysozyme on silica†
Physical Chemistry Chemical Physics ( IF 3.3 ) Pub Date : 2018-07-16 00:00:00 , DOI: 10.1039/c8cp03747g Nils Hildebrand 1, 2, 3, 4, 5 , Gang Wei 1, 2, 3, 4, 5 , Susan Köppen 1, 2, 3, 4, 5 , Lucio Colombi Ciacchi 1, 2, 3, 4, 5
Physical Chemistry Chemical Physics ( IF 3.3 ) Pub Date : 2018-07-16 00:00:00 , DOI: 10.1039/c8cp03747g Nils Hildebrand 1, 2, 3, 4, 5 , Gang Wei 1, 2, 3, 4, 5 , Susan Köppen 1, 2, 3, 4, 5 , Lucio Colombi Ciacchi 1, 2, 3, 4, 5
Affiliation
|
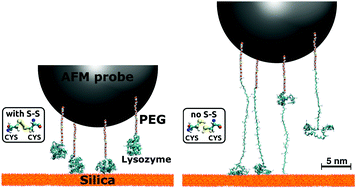
The force spectra of proteins detaching from oxide surfaces measured by atomic force microscopy (AFM) often present complex patterns of peaks, which are difficult to correlate with individual bond-breaking events at the atomic scale. In this work we rationalize experimental AFM force spectra of hen-egg-white lysozyme detaching from silica by means of all-atom steered molecular dynamics (SMD) simulations. In particular, we demonstrate that the native tertiary structure of lysozyme is preserved if, and only if, its four intramolecular disulfide bridges are intact. Otherwise, the protein pulled off the surface undergoes severe unfolding, which is well captured by SMD simulations in explicit solvent. Implicit solvent simulations, on the contrary, wrongly predict protein unfolding even in the presence of S–S bridges, due to the lack of additional structural stabilization provided by the water's hydrogen-bond network within and surrounding the protein. On the basis of our combined experimental and theoretical findings, we infer that the rugged force spectra characteristic of lysozyme/silica interfaces are not due to the successive breaking of internal disulfide bonds leading to partial unfolding events. Rather, they reflect the detachment of several molecules bound to the same AFM tip, each anchored to the surface via multiple hydrogen and ionic bonds.
中文翻译:

硅胶上溶菌酶的模拟和实验力谱†
通过原子力显微镜(AFM)测量的从氧化物表面分离的蛋白质的力谱通常呈现出复杂的峰模式,这些峰很难与原子级的单个键断裂事件相关联。在这项工作中,我们通过全原子导向的分子动力学(SMD)模拟来合理化从鸡蛋上分离出的鸡蛋类白色溶菌酶的实验AFM力谱。尤其是,我们证明,当且仅当其四个分子内二硫键完整时,溶菌酶的天然三级结构才能被保留。否则,从表面上拉下来的蛋白质会发生严重的折叠,这可以通过在显式溶剂中的SMD模拟很好地捕获。相反,即使在存在S–S桥的情况下,隐式溶剂模拟也错误地预测了蛋白质的展开,由于缺乏蛋白质内部和周围水的氢键网络提供的额外结构稳定性。根据我们结合实验和理论发现,我们推断溶菌酶/二氧化硅界面的崎force的力谱特征不是由于内部二硫键的连续断裂导致部分展开事件。而是,它们反映了绑定到同一AFM尖端的几个分子的分离,每个分子都锚定在表面上 我们推断溶菌酶/二氧化硅界面的崎force的力谱特征不是由于内部二硫键的连续断裂导致部分展开事件。相反,它们反映了绑定到同一AFM尖端的几个分子的分离,每个分子都锚定在表面 我们推断溶菌酶/二氧化硅界面的崎force的力谱特征不是由于内部二硫键的连续断裂导致部分展开事件。而是,它们反映了绑定到同一AFM尖端的几个分子的分离,每个分子都锚定在表面上通过多个氢和离子键。
更新日期:2018-07-16
中文翻译:

硅胶上溶菌酶的模拟和实验力谱†
通过原子力显微镜(AFM)测量的从氧化物表面分离的蛋白质的力谱通常呈现出复杂的峰模式,这些峰很难与原子级的单个键断裂事件相关联。在这项工作中,我们通过全原子导向的分子动力学(SMD)模拟来合理化从鸡蛋上分离出的鸡蛋类白色溶菌酶的实验AFM力谱。尤其是,我们证明,当且仅当其四个分子内二硫键完整时,溶菌酶的天然三级结构才能被保留。否则,从表面上拉下来的蛋白质会发生严重的折叠,这可以通过在显式溶剂中的SMD模拟很好地捕获。相反,即使在存在S–S桥的情况下,隐式溶剂模拟也错误地预测了蛋白质的展开,由于缺乏蛋白质内部和周围水的氢键网络提供的额外结构稳定性。根据我们结合实验和理论发现,我们推断溶菌酶/二氧化硅界面的崎force的力谱特征不是由于内部二硫键的连续断裂导致部分展开事件。而是,它们反映了绑定到同一AFM尖端的几个分子的分离,每个分子都锚定在表面上 我们推断溶菌酶/二氧化硅界面的崎force的力谱特征不是由于内部二硫键的连续断裂导致部分展开事件。相反,它们反映了绑定到同一AFM尖端的几个分子的分离,每个分子都锚定在表面 我们推断溶菌酶/二氧化硅界面的崎force的力谱特征不是由于内部二硫键的连续断裂导致部分展开事件。而是,它们反映了绑定到同一AFM尖端的几个分子的分离,每个分子都锚定在表面上通过多个氢和离子键。


























 京公网安备 11010802027423号
京公网安备 11010802027423号