当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Modeling Small-Molecule Reactivity Identifies Promiscuous Bioactive Compounds
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-07-10 00:00:00 , DOI: 10.1021/acs.jcim.8b00104 Matthew K Matlock 1 , Tyler B Hughes 1 , Jayme L Dahlin 2 , S Joshua Swamidass 1, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-07-10 00:00:00 , DOI: 10.1021/acs.jcim.8b00104 Matthew K Matlock 1 , Tyler B Hughes 1 , Jayme L Dahlin 2 , S Joshua Swamidass 1, 3
Affiliation
|
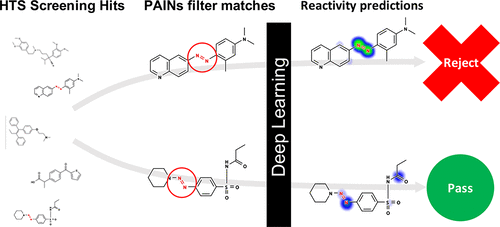
Scientists rely on high-throughput screening tools to identify promising small-molecule compounds for the development of biochemical probes and drugs. This study focuses on the identification of promiscuous bioactive compounds, which are compounds that appear active in many high-throughput screening experiments against diverse targets but are often false-positives which may not be easily developed into successful probes. These compounds can exhibit bioactivity due to nonspecific, intractable mechanisms of action and/or by interference with specific assay technology readouts. Such “frequent hitters” are now commonly identified using substructure filters, including pan assay interference compounds (PAINS). Herein, we show that mechanistic modeling of small-molecule reactivity using deep learning can improve upon PAINS filters when modeling promiscuous bioactivity in PubChem assays. Without training on high-throughput screening data, a deep learning model of small-molecule reactivity achieves a sensitivity and specificity of 18.5% and 95.5%, respectively, in identifying promiscuous bioactive compounds. This performance is similar to PAINS filters, which achieve a sensitivity of 20.3% at the same specificity. Importantly, such reactivity modeling is complementary to PAINS filters. When PAINS filters and reactivity models are combined, the resulting model outperforms either method alone, achieving a sensitivity of 24% at the same specificity. However, as a probabilistic model, the sensitivity and specificity of the deep learning model can be tuned by adjusting the threshold. Moreover, for a subset of PAINS filters, this reactivity model can help discriminate between promiscuous and nonpromiscuous bioactive compounds even among compounds matching those filters. Critically, the reactivity model provides mechanistic hypotheses for assay interference by predicting the precise atoms involved in compound reactivity. Overall, our analysis suggests that deep learning approaches to modeling promiscuous compound bioactivity may provide a complementary approach to current methods for identifying promiscuous compounds.
中文翻译:

模拟小分子反应性可识别混杂的生物活性化合物
科学家们依靠高通量筛选工具来识别有前途的小分子化合物,用于开发生化探针和药物。这项研究的重点是识别混杂的生物活性化合物,这些化合物在针对不同靶点的许多高通量筛选实验中表现出活性,但通常是假阳性,可能不容易开发成成功的探针。由于非特异性、难以处理的作用机制和/或干扰特定测定技术读数,这些化合物可能表现出生物活性。现在通常使用子结构过滤器来识别此类“频繁击球手”,包括泛分析干扰化合物 (PAINS)。在此处,我们表明,在 PubChem 分析中对混杂的生物活性进行建模时,使用深度学习对小分子反应性进行机械建模可以改进 PAINS 过滤器。在没有对高通量筛选数据进行训练的情况下,小分子反应性的深度学习模型在识别混杂的生物活性化合物方面的灵敏度和特异性分别为 18.5% 和 95.5%。这种性能类似于 PAINS 过滤器,在相同的特异性下实现了 20.3% 的灵敏度。重要的是,这种反应性建模是对 PAINS 过滤器的补充。当 PAINS 过滤器和反应性模型结合使用时,生成的模型优于单独使用任何一种方法,在相同特异性下实现 24% 的灵敏度。然而,作为一个概率模型,深度学习模型的敏感性和特异性可以通过调整阈值来调整。此外,对于 PAINS 过滤器的一个子集,这种反应性模型可以帮助区分混杂和非混杂的生物活性化合物,甚至在与这些过滤器匹配的化合物中也是如此。至关重要的是,反应性模型通过预测化合物反应性中涉及的精确原子,为测定干扰提供了机械假设。总体而言,我们的分析表明,建模混杂化合物生物活性的深度学习方法可以为当前识别混杂化合物的方法提供一种补充方法。反应性模型通过预测化合物反应性中涉及的精确原子,为测定干扰提供了机械假设。总体而言,我们的分析表明,建模混杂化合物生物活性的深度学习方法可以为当前识别混杂化合物的方法提供一种补充方法。反应性模型通过预测化合物反应性中涉及的精确原子,为测定干扰提供了机械假设。总体而言,我们的分析表明,建模混杂化合物生物活性的深度学习方法可以为当前识别混杂化合物的方法提供一种补充方法。
更新日期:2018-07-10
中文翻译:

模拟小分子反应性可识别混杂的生物活性化合物
科学家们依靠高通量筛选工具来识别有前途的小分子化合物,用于开发生化探针和药物。这项研究的重点是识别混杂的生物活性化合物,这些化合物在针对不同靶点的许多高通量筛选实验中表现出活性,但通常是假阳性,可能不容易开发成成功的探针。由于非特异性、难以处理的作用机制和/或干扰特定测定技术读数,这些化合物可能表现出生物活性。现在通常使用子结构过滤器来识别此类“频繁击球手”,包括泛分析干扰化合物 (PAINS)。在此处,我们表明,在 PubChem 分析中对混杂的生物活性进行建模时,使用深度学习对小分子反应性进行机械建模可以改进 PAINS 过滤器。在没有对高通量筛选数据进行训练的情况下,小分子反应性的深度学习模型在识别混杂的生物活性化合物方面的灵敏度和特异性分别为 18.5% 和 95.5%。这种性能类似于 PAINS 过滤器,在相同的特异性下实现了 20.3% 的灵敏度。重要的是,这种反应性建模是对 PAINS 过滤器的补充。当 PAINS 过滤器和反应性模型结合使用时,生成的模型优于单独使用任何一种方法,在相同特异性下实现 24% 的灵敏度。然而,作为一个概率模型,深度学习模型的敏感性和特异性可以通过调整阈值来调整。此外,对于 PAINS 过滤器的一个子集,这种反应性模型可以帮助区分混杂和非混杂的生物活性化合物,甚至在与这些过滤器匹配的化合物中也是如此。至关重要的是,反应性模型通过预测化合物反应性中涉及的精确原子,为测定干扰提供了机械假设。总体而言,我们的分析表明,建模混杂化合物生物活性的深度学习方法可以为当前识别混杂化合物的方法提供一种补充方法。反应性模型通过预测化合物反应性中涉及的精确原子,为测定干扰提供了机械假设。总体而言,我们的分析表明,建模混杂化合物生物活性的深度学习方法可以为当前识别混杂化合物的方法提供一种补充方法。反应性模型通过预测化合物反应性中涉及的精确原子,为测定干扰提供了机械假设。总体而言,我们的分析表明,建模混杂化合物生物活性的深度学习方法可以为当前识别混杂化合物的方法提供一种补充方法。


























 京公网安备 11010802027423号
京公网安备 11010802027423号